Transformers have shown promise in computational biology, but applying them to biological sequences could cause mispredictions in the downstream processes. A greater range of applications and more accurate downstream predictions are possible with three-dimensional (3D) molecule structures because they are more compatible with Transformer architecture and natural language processing models. This fundamental problem is addressed by Michigan State University researchers’ work using a topological transformer (TopoFormer). TopoFormer is a machine learning system that combines multiscale topology methods with natural language processing (NLP) to convert complex 3D protein-ligand complexes into sequences of topological invariants and homotopic forms at different spatial scales that are accepted by NLP. It outperforms standard algorithms and deep learning variations by embedding important physical, chemical, and biological relationships into topological sequences. AI-driven discovery is made possible by TopoFormer’s topological sequences, which can be derived from a variety of structural data sources supporting NLP models.
Introduction
Drug development has greatly benefited from conventional techniques in contemporary healthcare, such as molecular docking, free energy perturbation, and empirically based modeling. However, these processes are expensive and labor-intensive; a single prescription medication might cost billions of dollars and take more than ten years to complete. Due to these methodologies’ shortcomings—such as their inconsistent prediction capacities and computational intensity—therapeutic opportunities may be lost, or pharmacological efficacy may be incorrectly assessed. The application of cutting-edge techniques in drug discovery is nevertheless given top priority in modern healthcare despite these obstacles.
Bridging the Gap Between 3D Structures and Deep Learning Models
The Transformer framework and ChatGPT models are becoming popular choices in drug design because of their capacity to predict protein structures and decipher complex patterns. These models rely on large-scale pre-training and unlabeled data. Based on the achievements of bioinformatics and chemoinformatics, these models provide effective solutions when the availability of traditional labeled data is a constraint. However, because the Transformer framework ignores significant stereochemical linkages, its direct application to drug discovery raises concerns, especially for protein-ligand complex modeling. A key problem is adapting a paradigm for serialized language translations to the study of protein-ligand complexes, which naturally resist serialized representation. All things considered, the Transformer architecture provides interesting approaches to language processing and medication creation.
Understanding TopoFormer
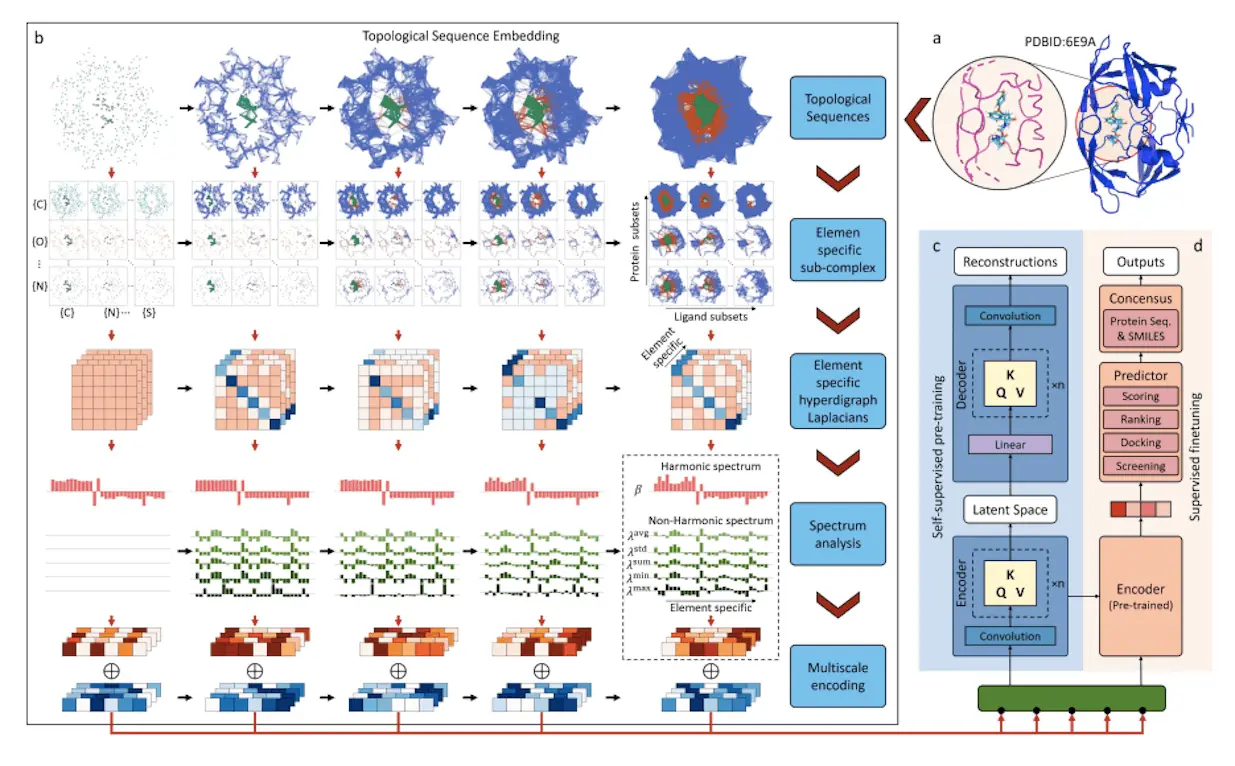
A topological transformer model called TopoFormer uses sophisticated mathematical models from combinatorial graph theory, differential geometry, and algebraic topology. Protein-ligand binding inherent physical, chemical, and biological interactions are captured by this multiscale model based on the persistent topological hyperdigraph Laplacian (PTHL). It is perfect for Transformers’ sequential architecture because it leverages PTHL’s multiscale topology and multiscale spectrum to translate complicated 3D protein-ligand complexes into 1D topological sequences. This novel combination of machine learning and topological insights transforms the knowledge of protein-ligand interactions. It redefines performance standards in drug development activities such as screening, docking, scoring, and ranking. The knowledge of protein-ligand interactions has changed dramatically as a result of TopoFormer’s sophisticated architecture.
Pre-training and Fine-tuning: Unlocking the Full Potential
The persistent topological hyperdigraph Laplacian (PTHL) and transformer are integrated for the first time in TopoFormer, a topological transformer model. Sequences of topological invariants, homotopic shape, and stereochemical evolution are produced by this model using 3D protein-ligand complexes as inputs. In a topological sequence acceptable to the transformer architecture, the PTHL technique successively embeds the homotopic shape, topological invariants, and physical, chemical, and biological interactions of these complexes. Pre-training the model on a variety of protein-ligand complexes helps it to understand the general features and subtleties of molecular interactions, such as stereochemical effects that are outside the scope of conventional molecular sequences. Deep learning is facilitated by further fine-tuning on particular datasets to guarantee that the output embeddings reflect the complex’s inherently complex interactions and represent the complex’s characteristics in touch with the entire dataset.
TopoFormer’s targeted analysis locates heavy ligand and protein atoms within a preset distance using a multiscale analysis. There are two models available: one with a cutoff of 20 Å and another with a cutoff of 12 Å. Using persistent topological hyperdigraph Laplacians (PTHLs), which embed element-specific PTHLs related to different physical, chemical, and biological interactions, the three-dimensional molecular structures are transformed into topological sequences. A series of embedding vectors are produced as a result of this multiscale investigation.
Conclusion
A wide range of unlabeled protein-ligand complexes are used by the self-supervised model TopoFormer to complete tasks. The Transformer encoder-decoder, which reconstructs the topological sequence embedding from its encoded version, is the central component of its architecture. This accuracy is measured by calculating the difference between the input and output embeddings. Following pre-training, the model moves to a supervised fine-tuning phase after familiarising itself with labeled protein-ligand complexes. TopoFormer is an excellent multitasker; it can handle docking, scoring, ranking, and screening. A number of topological transformer deep learning models (TF-DL) are started, each with a different random seed, to improve accuracy. Sequence-based models are also included to reduce biases. TopoFormer is a comprehensive model that combines deep learning and topological insights for protein-ligand interaction study.
Article Source: Published Abstract | Free Full Paper | Reference Article | Source codes and models are available publicly on GitHub
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.