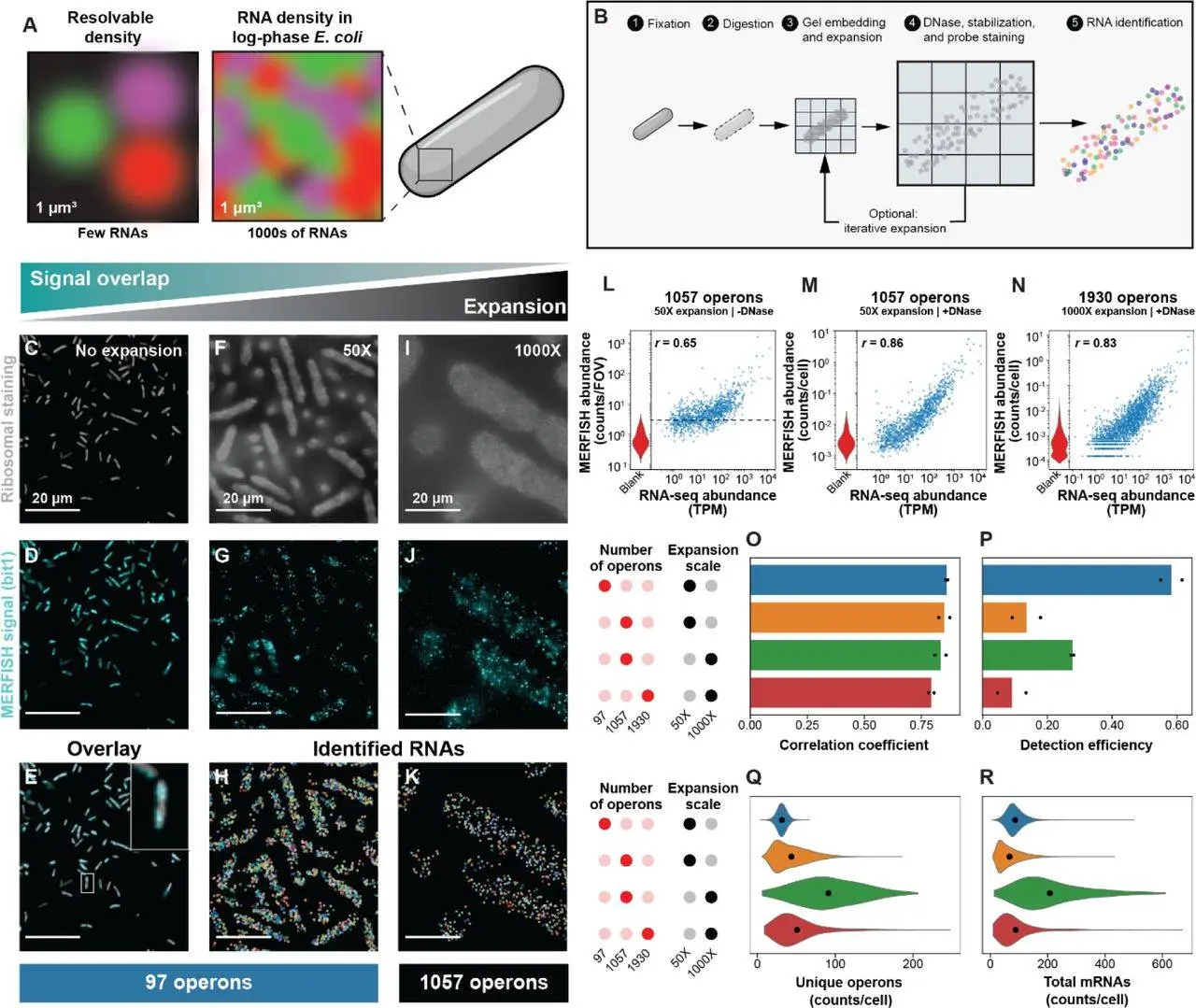
In one of the significant research studies, the researchers of Boston Children’s Hospital, Harvard Medical School, and Broad Institute of Harvard and MIT, Cambridge, presented a new technique known as bacterial-MERFISH to study bacterial behavior at the single-cell level. Packed with volumetric expansion coupled with multiplexed error-robust fluorescence in situ hybridization (MERFISH), it is a novel technique to perform high-throughput bacterial functional profiling in the context of operation in individual bacteria.
The Breakthrough: Bacterial-MERFISH
Initially, Bacterial-MERFISH was based on the integration of volumetric expansion and MERFISH, which allows for the high-throughput spatial analysis of several thousands of operons within a single bacterium. This technology solves the problem of high bacterial RNA density and is a flexible and high-efficiency solution for bacterial single-cell transcriptomics. Multi-faceted, Bacterial-MERFISH can be used to analyze single-cell behaviors, map spatial gene work, discern bacterial adaptability, meet microbial identity, strengthen antibiotic response research, and expand the knowledge base about host-symbiont interactions.
The Need for Multiplexing
Multiplexing is crucial in biology for the analysis of more than one target or parameter at a time in one experiment. In spatial transcriptomics, multiplexing enables the measurement of the expression of many genes or transcripts in individual cells or tissues. This capability is important when determining the genotype or phenotype of an organism because several genes or pathways are frequently implicated. Thus, obtaining such an all-around view by multiplexing helps to spot the patterns of gene expression, their relations, and the regulatory networks of the complex biological processes.
The Technology Behind Highly Multiplexed Spatial Transcriptomics
Of the existing techniques in highly multiplexed spatial transcriptomics, there is a technique called bacterial-MERFISH. This mind-boggling strategy helps in the integration of bacterially optimized expansion microscopy and a toolbox of multiplexed error-robust FISH. In this way, using these techniques, researchers can get single-cell characterization of a large fraction of the signal transcriptome, with thousands of operons being profiled with a high rate of detection and accuracy and within a thousand-fold throughput in bacterial cells. It solves the problem of relatively high bacterial RNA density and provides a spatially resolved transcriptome-scale analysis of gene expression within individual bacteria, which is a general and high-performance platform for bacterial SC personally.
Key Findings
Spatial Variation in B. theta Operons: The study disclosed that B. theta operons showed a substantial spatial difference in expression in cross-sections of the colonic crypts. The distribution patterns of some mRNAs included expression in the lumen of the crypts, while others were stronger in the region closer to the mucus layer.
Quantification of Spatial Variation: Further, to assure the validity of data, measurements were also taken in several colonic slices from different mice, which helped the researchers to assure the relation between two fixation methods and gene expression. They averaged gene expression over small spatial areas that encompassed about five B. theta cells.
Potential of Bacterial-MERFISH: These results highlight the capability of the bacterial-MERFISH in investigating single-cell variations in numerous biology and spatial settings. This approach has given hope of exposing some of the secrets bacteria have in today’s world.
Applications
It is possible to list several application areas of highly multiplexed spatial transcriptomics in bacterial research. This technology allows researchers to:
Study Single-Cell Behaviors: Get an understanding of the decision-making process, behavior, and response within bacterial populating cells.
Explore Spatial Gene Expression: Learn about the differences in gene regulation on one or multiple bacterial cells or groups of cells.
Investigate Bacterial Adaptation: Identify the modus operandi of bacteria as it pertains to the diversification of specific niches as well as varied environments.
Characterize Microbial Communities: Investigate the taxonomy of metabolic genes and RNA sequences involved in gene expression in dense microbial communities and biofilms.
Enhance Antibiotic Response Studies: Enhance the knowledge about antibiotic responses and persistence in bacterial systems.
Deepen Host-Microbe Interaction Understanding: Analyze the changes in gene activity during host-microbe interactions, which can be mutualistic or pathogenic.
Challenges
The complexity of Spatial Data: Processing ‘omics’ data from bacterial cells is often problematic because the spatial organization of cells contains multivariate gene-expression profiles.
Technical Optimization: Extreme care should be taken to ensure that the accuracy and throughput of highly multiplexed spatial transcriptomics methods are standardized across the broad range of bacterial species and environments.
Interpretation of Spatial Heterogeneity: Having insight into the functional consequences of among-population differences in gene distribution space.
Future Directions
Method Refinement: Further improvements include optimizing spatial transcriptomics techniques to improve spatial resolution, sensitivity, and time efficiency.
Integration with Multi-Omics: The need for assimilation of information derived from various omics approaches complemented with spatial transcriptomics data to offer a rich appreciation of bacterial functions.
Exploration of Novel Environments: Applying SPT to investigate bacterial behaviors in new environments/milieu or under different conditions.
Machine Learning Applications: Applying its features in the statistical optimization of raw and layered spatial transcriptomics data in bacteria.
Clinical and Environmental Applications: Appointing spatial transcriptomics for clinical purposes and using the environmental microbiome for various purposes.
Conclusion
The study is a major advancement in knowledge of bacteria’s conduct at a cellular level. Comparing the spatial context to gene expression, bacterial-MERFISH is an invaluable approach to understanding the bacterial population’s multifaceted activity better. More and more bacterial transcriptomics are being studied daily; however, research like this creates the foundation for further advancements in various fields, including microbiology.
Article Source: Reference Paper
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.