Protein structure is important and should be studied to understand many biological processes and disease progressions. There are many advancements using machine learning techniques, but accurate protein sequencing remains a challenge in the field. Protein sequencing typically depends on mass spectrometry (MS), and they face limitations in accurately identifying all the amino acids. Researchers from the University of Texas at Arlington and UCLA Health try to simulate the real world by masking amino acids that are hard to experimentally identify within protein sequences and then fine-tune ProtBert to predict the masked residues showing structural assessments done using AlphaFold and TM-score validation.
Introduction: Peptides in Brief
Peptides are small chains of amino acids joined with peptide bonds and are active components of many biological processes. They fill in the size gap between small molecules and big proteins in many physiological functions. Whereas proteins are in long chains of amino acids, peptides are generally in 2 to 50 amino acid chains in length. Though relatively very small, peptides are found to be engaged in nearly every process in life and are essential for life.
The fact is that peptides are structurally diverse; therefore, they can cause a great diversity of functions. What appreciates the structure and function of peptides is the sequence of amino acids that participate in the peptide. Hence, small changes in sequence grossly change peptide activity, leading to differences in biological outcomes. This, again, is an important fact that makes peptides invaluable tools within sciences and medicine.
Challenge of Sequencing Proteins
Traditional protein sequencing techniques have, for many years, formed the basis of proteomics science. These techniques, however, are afflicted with numerous limitations. For example, MS may not identify all the amino acids in a sequence, especially where complex proteins are involved. Edman degradation is tough; therefore, it can process only relatively short sequences. Most of the time, these limitations lead to partially known sequences, preventing the construction of a comprehensive proteome analysis.
Recently, both the fields of click chemistry and bioorthogonal chemistry have made attempts to overcome some of these challenges in identifying specific amino acids and their locations. Currently, these techniques are limited by the quantity of precisely identifiable amino acids, thus producing sequences that contain gaps.
A New Approach Using Language Models for Proteins
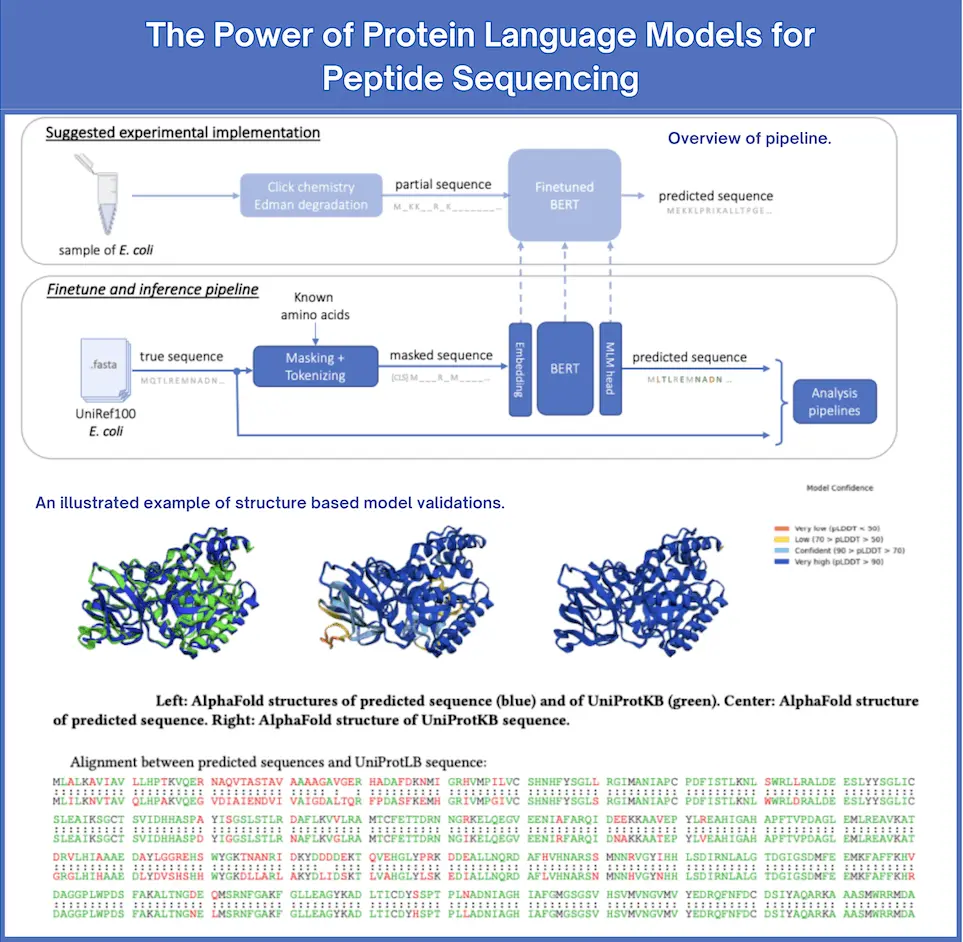
The authors suggest a method to simulate such partial sequencing data, masking the amino acids that are experimentally hard to identify, mimicking real-world limitations to sequencing. After this, they use ProtBERT, a transformer-based language model that predicts these masked residues, providing a probabilistic reconstruction of the full protein sequence.
Model Fine-Tuning and Evaluation
The researchers fine-tuned a pre-trained language model, ProtBERT, to suit their needs in protein sequencing. Generally, ProtBERT was developed to analyze protein sequences and was fine-tuned here to predict missing amino acids in partially sequenced proteins. The model has been trained on protein sequences from three Escherichia species: E. coli, E. albertii, and E. fergusonii.
During training, some of the sequences are masked- that is, the model is trained on input sequences where certain amino acids are replaced. This simulates limitations in experimental techniques. The goal was to predict what masked amino acids might be, essentially filling in the gaps in the sequence. The fine-tuned model showed remarkable accuracy; per-amino acid accuracy reached as high as 90.5 percent on the basis of only four known amino acids: K, C, Y, and M.
Model Predictions Validation
They used the state-of-the-art tool AlphaFold to predict protein structures from their amino acid sequences and validate the biological relevance of the model’s predictions. Comparisons of the predicted structures against actual structures of proteins confirmed that model predictions were not only accurate but, indeed, biologically meaningful.
This was further validated through the tests for generalizability across different species. Although trained on data from specific Escherichia species, it demonstrated high accuracy when applied to other species, thus proving its potential for applications in proteomics at large.
Possible Future Directions
This new concept is an exciting development for us! It is the future of proteomics. This means that when traditional methods have their limitations, it would be a good way to combine experimental data with computational predictions to offer complete and accurate protein sequencing. Such a strategy might accelerate progress in proteomics and structural biology toward the eventual deciphering of mechanisms underlying protein structure and function.
Moreover, the integration of the protein language models with experimental data opens up several future paths for evolutionary analysis. Since the model generalizes across species, one can use such a model to study evolutionary relationships between proteins, a task from which beneficial insights into mechanisms of evolution at the molecular level could be gained. Perhaps the best futuristic idea is- liquid biopsies. Protein sequencing by language models will have very important implications for constructing liquid biopsies. The researchers have identified urine as the “gold standard” for a liquid biopsy since they are safe, non-invasive, and easier to work with than blood plasma.
Conclusion
This new approach proposed by these researchers is a huge enhancement to the state of the field in protein sequencing. Using the powers of large language models, the researchers worked out a way to accurately predict complete protein sequences from partial experimental data. This approach is not only able to overcome the limitations linked with conventional sequencing methods but also opens a new way to do research in proteomics and the study of protein structure. While this is an approach to continuous refinement and expansion, it can be game-changing in the way we study proteins and know about their biology!
Article Source: Reference Paper | GitHub
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Neermita Bhattacharya is a consulting Scientific Content Writing Intern at CBIRT. She is pursuing B.Tech in computer science from IIT Jodhpur. She has a niche interest in the amalgamation of biological concepts and computer science and wishes to pursue higher studies in related fields. She has quite a bunch of hobbies- swimming, dancing ballet, playing the violin, guitar, ukulele, singing, drawing and painting, reading novels, playing indie videogames and writing short stories. She is excited to delve deeper into the fields of bioinformatics, genetics and computational biology and possibly help the world through research!