The entire process of drug discovery costs more than a billion dollars and takes over a decade to complete. This is an expensive and time-consuming process. However, to help improve efficiency, DNA Encoded Library (DEL) technology has been introduced, which enables researchers to screen billions of small molecules for possible drug candidates. The most challenging aspect of running DEL experiments is analyzing the massive amount of data generated. There are a few proprietary tools available to assist with this, and especially for smaller research groups, these remote tools often remain unreachable.
In a recent study, researchers from the Center for Integrative Chemical Biology and Drug Discovery at the Eshelman School of Pharmacy, University of North Carolina, introduced DELi—a groundbreaking open-source platform for DEL informatics. DELi aims to democratize DEL technology by offering a free and accessible tool for library design, sequencing data decoding, and hit analysis.
Why DELi Matters
Data analysis remains a primary bottleneck despite the rapid growth of DEL’s adoption. The selection of DEL’s results involves an analysis whose intricacy requires knowledge of chemistry, sequencing, and data science. Proprietary approaches do not offer sufficient granularity and are too inaccessible for most academic and small biotech companies. DELi fills this gap by providing a fully automated and open-source solution for DEL informatics.
Key Features of DELi
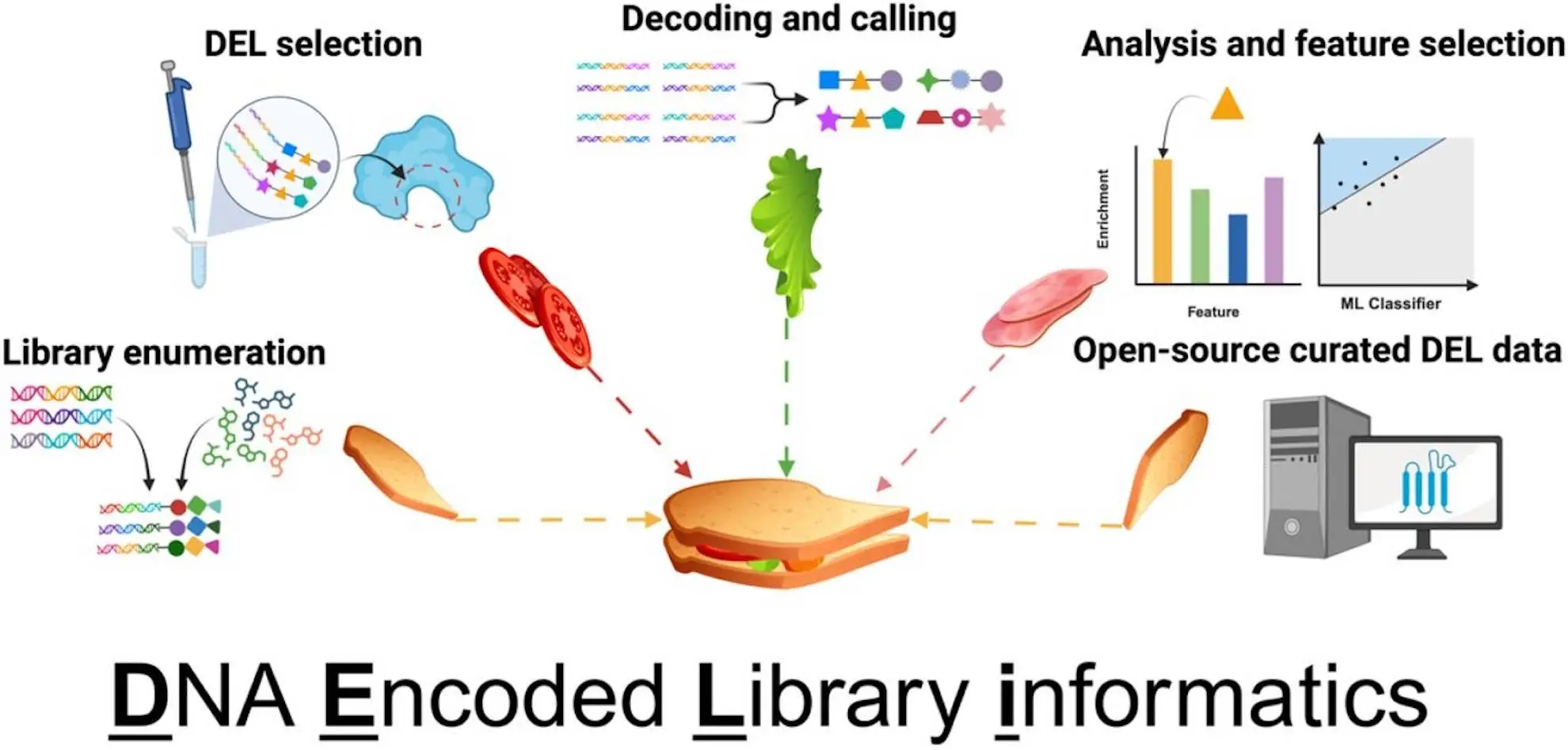
DELi is implemented in Python and follows good software engineering practices. Like most systems, DELi contains various components that automate the DEL workflow. From barcode design to compound selection:
- Barcode Design: DELi supports the creation of error-correcting DNA barcodes using Hamming codes, which help correct sequencing errors and improve data accuracy.
- Library Enumeration: This module algorithmically constructs all possible compounds in a DEL, which may reach billions, based on user-specified building blocks.
- Barcode Decoding: DELi translates raw sequencing reads to compound counts in a fast fashion by using a semi-global alignment algorithm.
- DEL Analysis: The statistical analysis module applies methods of information theory such as normalized sequence count (NSC) and maximum likelihood enrichment ratio (MLE) alongside machine learning to boost promising hit compound detections.
- Machine Learning Integration: DELi incorporates predictive modeling techniques, such as graph convolutional networks and random forest classifiers, to enhance hit identification and optimization.
DELi in Action: A Case Study with BRD4
To test DELi’s performance, the researchers developed a custom DNA-Encoded Library called UNC-DEL006 featuring benzimidazole-based compounds. They did selection experiments with Bromodomain Bound Protein 4 (BRD4), a prominent target for cancer. DELi accurately detected UNC 002-080 as a leading compound with nanomolar binding affinity. By contrast, a molecule (UNC 002-083) structurally related was not prioritized by DELi, which was found to show no measurable binding. This case illustrates DELi’s capability of distinguishing the most relevant drug candidates from a pool of false positives.
Conclusion
DELii marks, perhaps, the first step towards making workable system DEL technology more accessible and efficient. With proprietary software no longer obstructing the way, DELi gives researchers the freedom to optimize the use of DNA-encoded libraries in drug discovery.
The researchers intend to move DELi to open access in the hope that it will inspire additional development and creativity in the area of DEL technology. Programs such as DELi, which are open source, can be utilized by researchers in any part of the world to create new DEL techniques, apply modern artificial intelligence, and even revolutionize processes of developing drugs. The researchers are also preparing user-friendly extensions, including more advanced interfaces to enrich DEL automation and more powerful machine learning components.
As the field continues to evolve, it is evident that open-source projects such as DELi will be at the forefront of shaping the destiny of computational chemistry and more medicinal initiatives.
Article Source: Reference Paper.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.