Scientists from GAO Caixia’s team at the Chinese Academy of Sciences’ Institute of Genetics and Developmental Biology (IGDB) with collaborators have introduced Engineered Plant Prime Editor, a more efficient and enhanced plant prime editing system (ePPE). The new editing approach is projected to broaden the scope of prime editing’s application in agricultural breeding, crop improvement, and more.
Prime editing is a powerful genome editing method; however, it has low efficiency in editing. It is a recently described precision genome editing tool that can generate precise base substitutions, deletions, or insertions of DNA bases at a specific target place in the genome of living cells.
Unlike prior studies that concentrated on improving prime editing efficiency by primarily refining the pegRNA, this time, the researchers focused on the modification of protein components in the prime editor. The findings were reported in the journal Nature Biotechnology in the last week of March.
The first inception and optimization of using prime editing were successfully led by GAO’s team in 2020 when the team explored the rice and wheat through the fusion of various reverse transcriptase enzymes, suitable experimental temperatures, pegRNA expression methods, primer binding site sequence, and reverse transcriptase (RT) template sequence lengths and other conditions. Nonetheless, prime editors for use in plants had a low editing efficiency and were highly target-dependent.
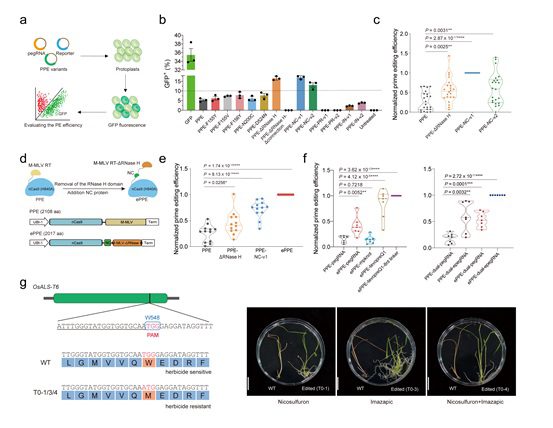
Image Source: https://english.cas.cn/newsroom/research_news/life/202203/t20220323_302939.shtml
The Moloney–murine leukemia virus reverse transcriptase was modified by deleting the ribonuclease H domain and replacing it with a viral nucleocapsid protein that acts as a nucleic acid chaperone. Each alteration increased prime editing efficiency in plant cells by 1.8–3.4-fold.
The researchers also developed the improved engineered plant prime editor, or ePPE, by combining both methodologies. When evaluating different editing types, such as base substitutions, small fragment insertions, deletions, and insertions or deletions of larger fragments, ePPE can increase editing efficiencies by an average of 5.8-fold compared to the original PPE without increasing the occurrence of off-target editing events or editing by-products.
The effectiveness of the prime editing system for generating precise editing events at endogenous genes can be further improved by combining the newly developed ePPE with the dual-pegRNA strategy previously reported by GAO’s team and the epegRNA (Engineered pegRNA) strategy developed by David Liu’s group in 2021.
The researchers successfully employed the ePPE method to create new rice plants that are herbicide-resistant to imazamox nicotinic acid and nicosulfuron.
The new ePPE system developed can dramatically improve prime editing efficiencies in plants by modifying the protein components of PPE, and the coupling of ePPE with pegRNA optimization methodologies can further improve editing efficiency across a number of cell types.
Story & Image Source: Zong, Y., Liu, Y., Xue, C. et al. An engineered prime editor with enhanced editing efficiency in plants. Nat Biotechnol (2022). https://doi.org/10.1038/s41587-022-01254-w https://english.cas.cn/newsroom/research_news/life/202203/t20220323_302939.shtml
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar