The National Institutes of Health's discovery could lead to new approaches to tackle future SARS-CoV-2 variants.
According to a study by experts at the National Institutes of Health, unlike other SARS-CoV-2 variants, the Delta variant can bind to copies of itself, generating larger aggregations, or clumps, of virus particles. According to the researchers, this connecting trait may have contributed to the Delta variant’s propensity to spread faster than other variants before.
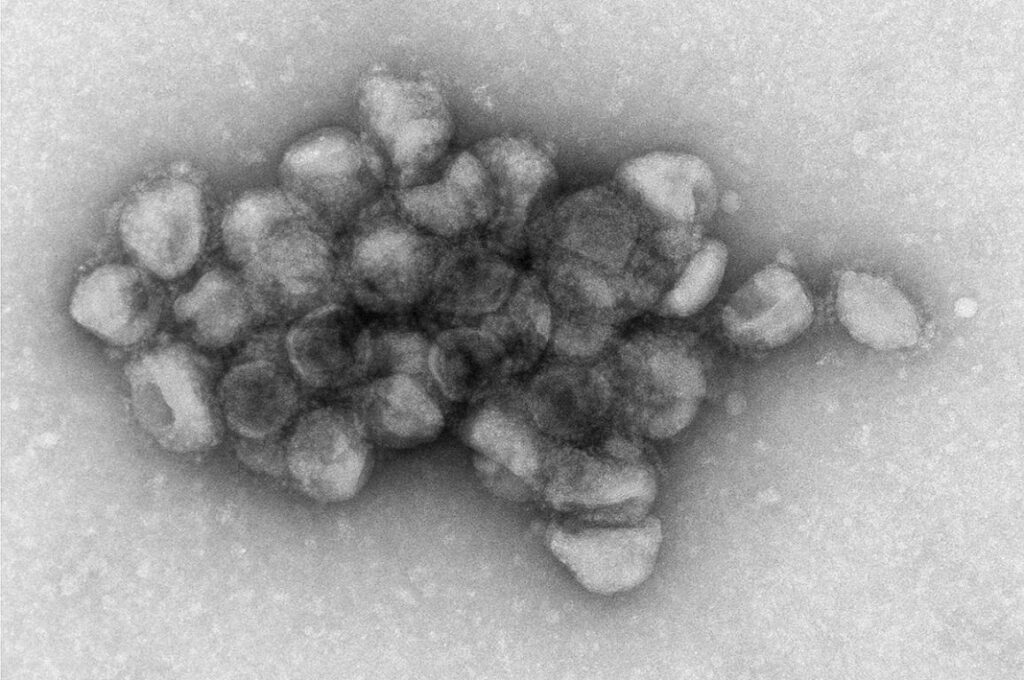
Image Source: https://www.nih.gov/news-events/news-releases/unique-binding-delta-variant-may-explain-high-transmissibility
Jennifer D. Petersen, Ph.D. of the NIH’s Eunice Kennedy Shriver National Institute of Child Health and Human Development, and colleagues conducted the research.
As the severe acute respiratory syndrome coronavirus 2, SARS-CoV-2, spreads worldwide, new genetic variants emerge, each with the ability to elude acquired immunity and re-ignite the COVID-19 pandemic.
Individuals infected with the SARS-CoV-2 Delta variant, lineage B.1.617.2, have a faster initial infection and a higher viral load than those infected with previous variants, and pseudotyped viral particles bearing the SARS-CoV-2 Delta variant spike protein infect target cells faster than those bearing other SARS-CoV-2 variant spikes.
The Delta spike protein allows the virus to attach to cells and begin infecting them. The researchers detected this effect in laboratory trials using pseudotyped Delta particles, which are leukemia viruses from mice stripped of disease-causing genes but designed to have the spike protein on their surface. According to an earlier study, the researchers saw the spike proteins interacting with one another to create aggregations, which boosts the likelihood of viral propagation.
Deep inside the aggregation, viruses are protected from drying out, antiviral medications, and the host’s immune system. Furthermore, large viral aggregations have the ability to bring a larger number of viruses into contact with target cells, boosting the likelihood of infection. According to the scientists, future research is needed to confirm whether the SARS-CoV-2 Delta variant can form aggregations comparable to the Delta pseudo particles.
According to the researchers, engineered particles of the Omicron variety, which has subsequently superseded the Delta variant, do not form aggregations.
Viral aggregation can improve fitness by sheltering virions from environmental risks and allowing for the release of numerous viral genomes at the same time, known as collective infection. In rare cases, a joint infection may favor the initial infection. The size and number of virions per aggregate are likely critical factors in boosting infectivity—too many virions in an aggregate and the benefit of collective infection is lost; too few virions in an aggregate and the benefit of collective infection is lost.
The unique property of the Delta spike to aggregate pseudotyped viral particles (PVs) may underlie the faster infection by Delta PVs. Furthermore, spike-mediated aggregation could be part of the molecular mechanism by which Delta variant SARS-CoV-2 achieves increased transmissibility and faster disease with a higher viral load.
The Delta spike’s unique ability to aggregate PVs could explain why Delta PVs infect faster. Spike-mediated aggregation could also be a part of the molecular process by which Delta variant SARS-CoV-2 achieves greater transmissibility and quicker infection with a higher viral load.
However, the researchers believe that understanding how Delta aggregates—and developing treatments that can target viruses inside the aggregation—could be valuable if a future variant capable of aggregation emerges.
Story Source: Petersen JD, Lu J, Fitzgerald W, Zhou F, Blank PS, Matthies D, Zimmerberg J. Unique Aggregation of Retroviral Particles Pseudotyped with the Delta Variant SARS-CoV-2 Spike Protein. Viruses. 2022; 14(5):1024. https://doi.org/10.3390/v14051024 https://www.nih.gov/news-events/news-releases/unique-binding-delta-variant-may-explain-high-transmissibility
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar