Protein structure prediction is a pretty big challenge in the field of bioinformatics as it depends on a multitude of factors. Imagine trying to understand a random paragraph in a novel from just its sentences, without any context of the chapters before it. In the same way, the primary structure (sequence of amino acids) isn’t enough to describe the entire functioning of the protein. Accurately predicting the folded structure of a protein de novo is very laborious. Hence, protein design and engineering to optimize structures for certain purposes is strenuous, too. However, there have been substantial advances over the past few years, which can broadly be attributed to machine learning (ML). These advances have led to the production of more stable, catalytically active, and specific proteins with huge applications for biotechnology, medicine, and sustainability. However, many of these ML tools are proprietary or require a set of skills that is not easy to obtain freely, thus limiting their overall applicability. Researchers from the Technical University of Denmark have developed ProteusAI, the new, open-source, user-friendly platform meant to democratize access to machine learning-guided protein design and engineering.
Protein Engineering and Machine Learning
Protein engineering (PE) is the designing or optimization of proteins to create novel therapeutics, enzymes, or environmentally friendly solutions. Traditional approaches, such as directed evolution (DE), have many rounds of gene mutagenesis and subsequent selection. DE has proven useful but problematic indeed: slow, expensive, and inefficient; for the most part, this is because the search space for protein sequences is huge, and it’s not possible to predict which mutations will confer improved functionality. In addition, mutations may lead to trade-offs between such important protein properties as stability and activity, adding yet another layer of complexity to the process.
Such challenges call for the advancement and power of machine learning. These ML models can predict the effects of mutations; they can thus optimize protein functions more accurately and speedily than traditional approaches. They can process vast datasets of protein sequences to identify patterns, make predictions, and guide the experimentalists in selecting the most promising candidates for further study.
Despite the great promise that ML holds for PE, most current tools are too sophisticated for anyone except experts or are unavailable as open-source solutions. To bridge that gap, ProteusAI was developed to offer an ML platform that is accessible to users ranging from academic researchers to students, teachers, and the common man.
What is ProteusAI?
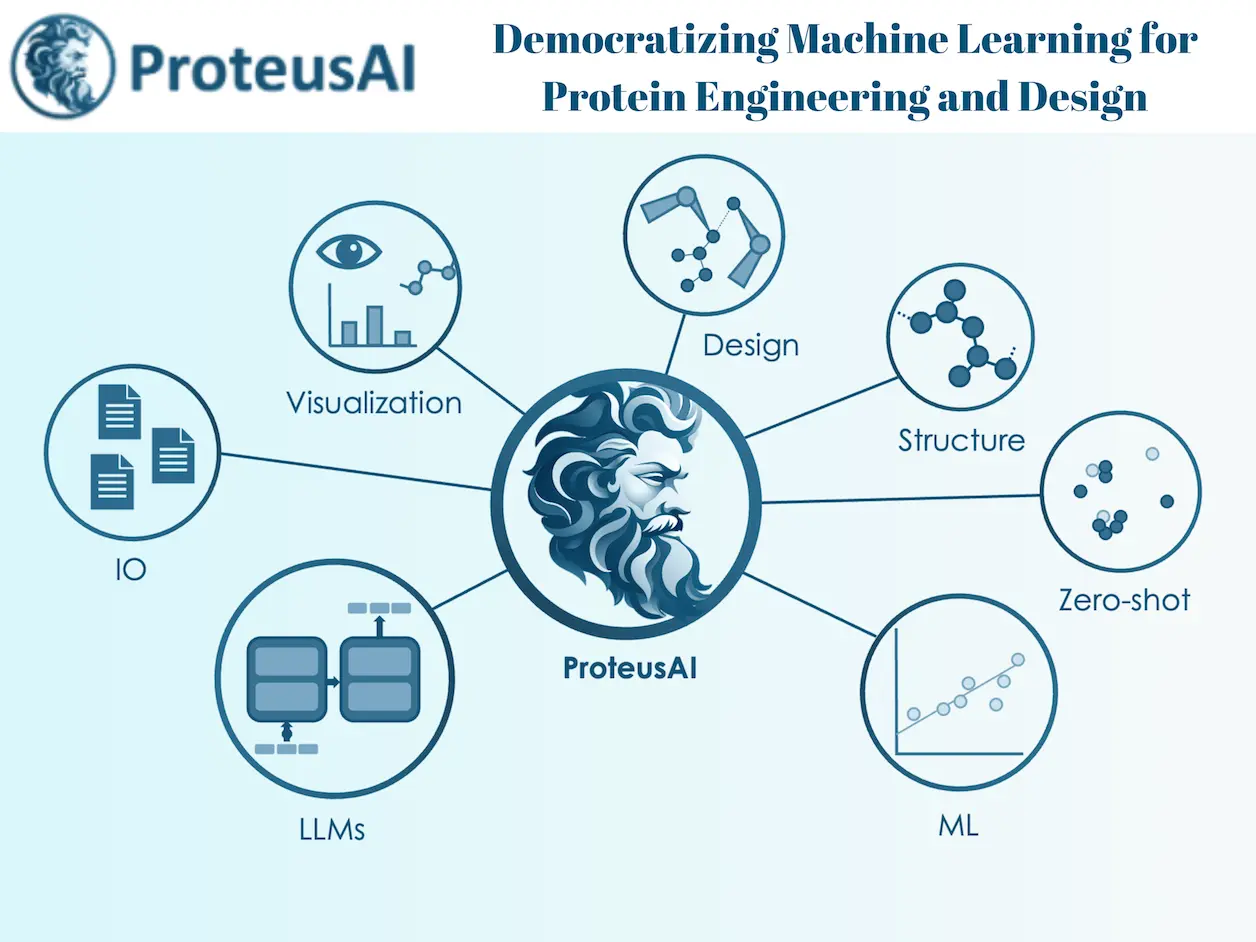
ProteusAI is an open-source platform for supporting the needs of protein engineers in the design-build-test-learn (DBTL) cycle. The tools provided can be applied to any stage of the process of discovering new proteins to optimize stability and activity through ML-guided directed evolution.
These comprise a web-based application accessible through a Python package or open-source code on GitHub. Such openness allows users to use the platform irrespective of their technical background and whether they require an easily accessible graphical interface or direct integration with workflows based on Python.
Modules in ProteusAI
The ProteusAI has a few central modules:
Protein Discovery Module
This module allows the researcher to discover functional proteins even from large sequence databases, where experimental annotations may not be provided. Combining protein language models with a vast amount of evolutionary information, ProteusAI predicts how functional similarities are shared between sequences and clusters them according to these similarities so that promising candidates are selected for follow-up experimental validation.
It usually involves stabilizing, re-solubilizing, or overexpressing a protein without impairing its function. ProteusAI uses inverse folding algorithms for structure-based design to suggest protein sequences that enhance these properties.
Module for Zero-Shot Prediction
Of course, one of the big challenges in the engineering of proteins is how to design the primary mutant library that launches the optimization process. The zero-shot (ZS) module exploits protein language models (PLMs) to generate predictions about the influence of mutations on function without needing prior experimental data. This is why it is called zero-shot; labeled training data is not required. The work described here led to the enrichment of mutant libraries with potentially advantageous variants, thereby jump-starting the optimization process and reducing the number of rounds needed to achieve desired outcomes.
Machine Learning-Guided Directed Evolution (MLDE) Module
The MLDE module enables the iterative improvement of protein properties by using experimental data for updates. Using experimental results for the training of ML models that, in turn, use a BO strategy (a probabilistic surrogate model (SM) predicts fitness values and uncertainties, and an acquisition function (AF) weighs the prediction and uncertainty to prioritize novel sequences for further testing) guides ProteusAI towards optimization of functions such as catalytic activity or binding affinity with a minimal number of experiments.
Benchmarking and Performance
ProteusAI was very effective at finding the best-performing variants in an optimal number of experimental rounds. It was specifically proven that, when combined with ESM-2 representations and the Expected Improvement acquisition function, the Random Forest (RF) models on this platform are the leading ones in finding the best mutants.
One important takeaway of the benchmarking study is that RF models perform well even when data is limited. This characteristic is important for early-stage research projects since generating large amounts of data can be costly and time-consuming.
Open Source and Future Developments
One of the biggest advantages of ProteusAI is that it can be built based on an open-source model, so people in the community can drive development and customize it. By making the system free, the developers hope to generate collaboration among researchers and speed up the pace of developing new perspectives in protein engineering.
Conclusion
ProteusAI is a landmark for the democratization of protein engineering! It would provide the researcher with an easy-to-use, open-source platform integrating the newest machine learning tools, which could be used to design and optimize proteins, empowered to design and optimize proteins more efficiently than ever before. ProteusAI is sure to become the lifesaver of protein engineering for academia and industry.
Article Source: Reference Paper | ProteusAI is available through http://proteusai.bio/ | Code is available on GitHub.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Neermita Bhattacharya is a consulting Scientific Content Writing Intern at CBIRT. She is pursuing B.Tech in computer science from IIT Jodhpur. She has a niche interest in the amalgamation of biological concepts and computer science and wishes to pursue higher studies in related fields. She has quite a bunch of hobbies- swimming, dancing ballet, playing the violin, guitar, ukulele, singing, drawing and painting, reading novels, playing indie videogames and writing short stories. She is excited to delve deeper into the fields of bioinformatics, genetics and computational biology and possibly help the world through research!