Scientists at the Center for Integrative Medical Sciences Yokohama, Japan, designed a platform-independent application to run UniverSC, a wrapper for the 10X Genomics CellRanger program that works with different single-cell technologies, via a graphical user interface, operable on macOS, Windows, and Linux Ubuntu, eliminating data processing for single-cell RNAseq (scRNAseq) analysis for researchers who are naive to bioinformatics. It is envisaged that UniverSC will make single-cell analysis reliable and foolproof to utilize, revolutionizing scRNAseq technology.
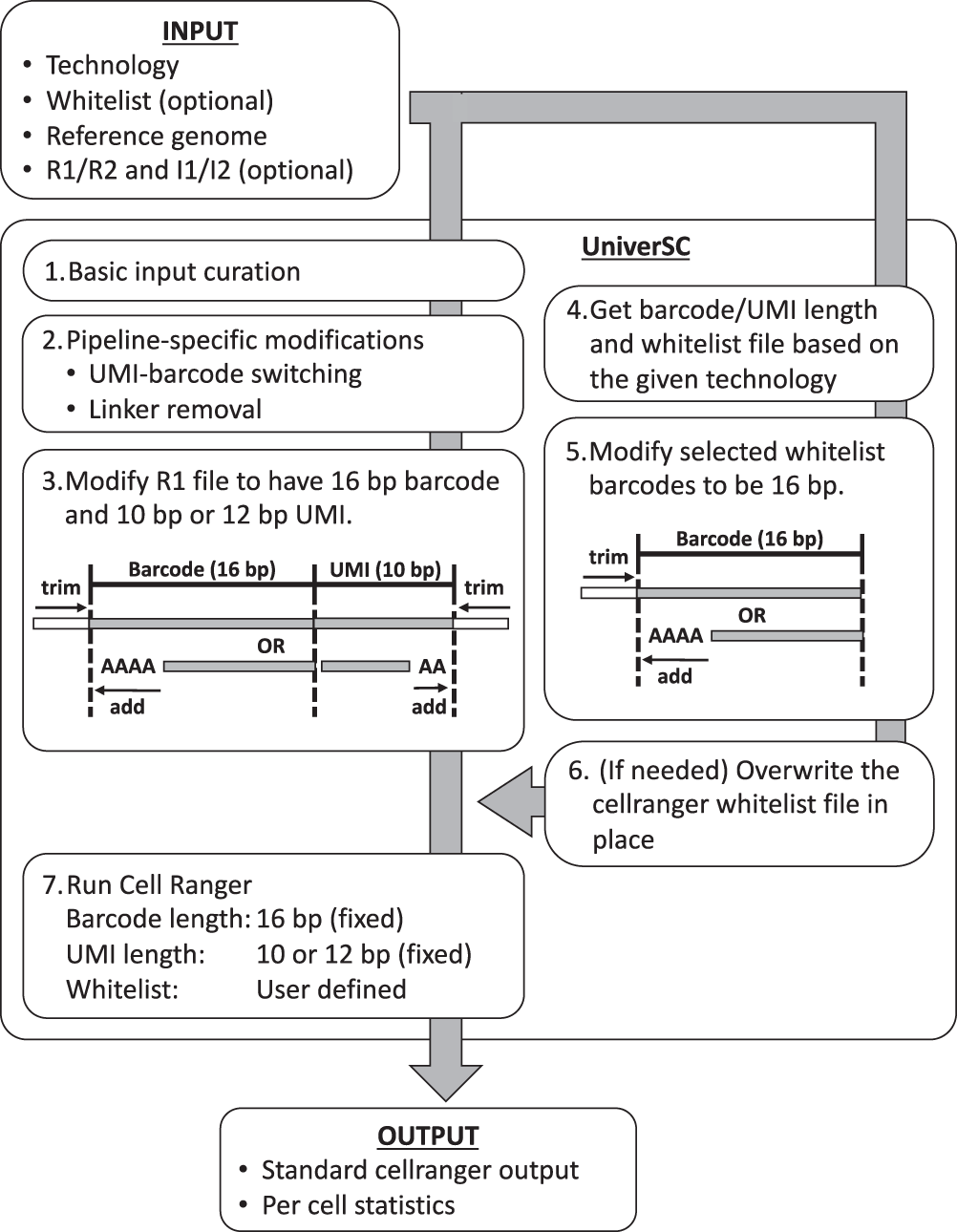
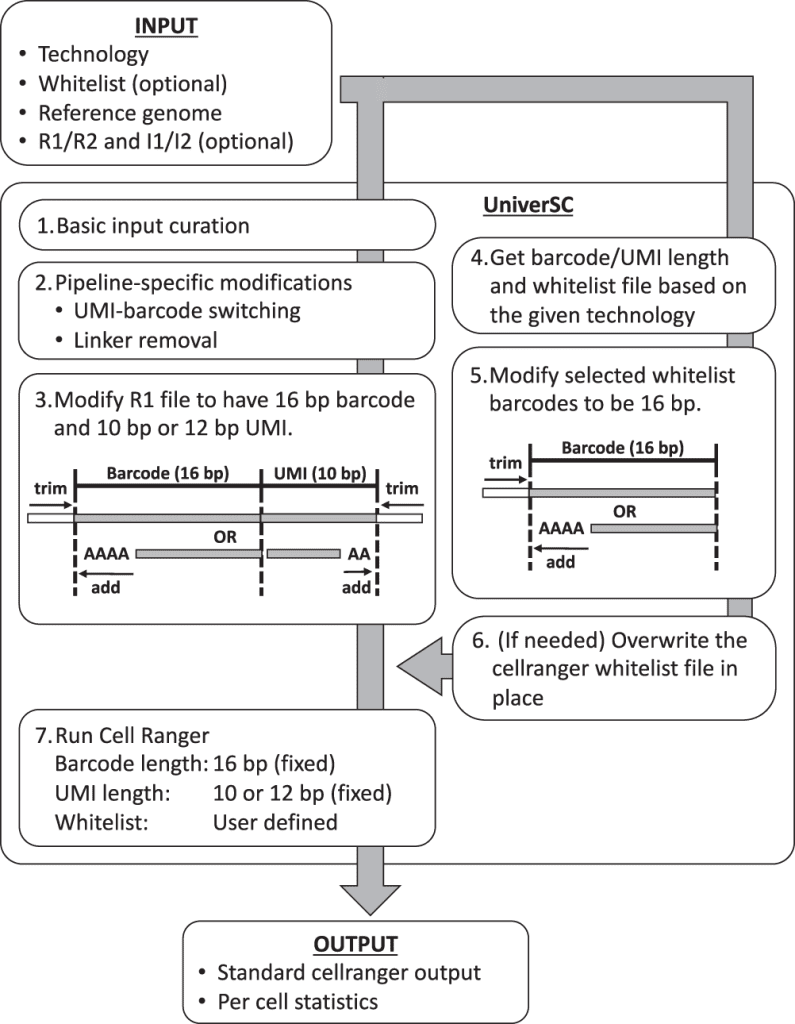
UniverSC Work-flow
- UniverSC first runs a basic input curation on a given pair of FASTQ files (R1 and R2), a genome reference.
- Next, the curated input files are adjusted for pipeline-specific modification by aligning curated input files.
- Reformatting to match the expected barcode and UMI lengths.
- Determination of barcode whitelist suited for the technology
- Modification of whitelist barcodes to 16 bp.
- The whitelist is replaced if the chosen whitelist differs from the one already in use for Cell Ranger.
- Generating standard output by processing modified sample data using Cell Ranger against the modified whitelist along with a summary file with per-cell statistics.
To be compatible with CellRanger, UniverSC alters the cell barcode and Unique Molecular Index (UMI), allowing users to create gene expression matrices using a range of single-cell technologies. The authors compare the results of UniverSC against those of existing pipelines for 10X genomics, Drop-seq, and ICELL8 technologies before showcasing UniverSC on those datasets.
Single-cell RNA-sequencing analysis to measure the RNA molecules in individual cells can gather a lot of data from each experiment and has therefore grown tremendously. Single-cell sequencing offers the ability to reveal cellular population heterogeneity at the genomic, epigenomic, and transcriptome levels, as well as changes at these levels.
Research of cellular heterogeneity has recently exploded, spurred by single-cell genomics tools. Cell throughput has risen over time, and modern scRNAseq methods, some commercially accessible, may regularly output data for thousands to hundreds of thousands of cells in a single experiment. Researchers may now use scRNAseq in a variety of tissues and whole organisms thanks to this improvement in throughput. As the technology advances, it is anticipated that scRNAseq will improve in accuracy, dependability, and cost per cell, making it practical for a variety of studies. The capacity of biologists to interpret the data when it is generated, however, is still a constraint.
With the command-line interface, UniverSC, which is publicly accessible on GitHub and DockerHub, may be used on any Unix-based machine. It may also be operated on Ubuntu, MacOS, and Windows using a graphical user interface (GUI), thus, installing or configuring different pipelines for each platform is no longer necessary.
Cross-platform single-cell data integration by UniverSC.
A typical approach for many scRNAseq methods requires catching individual cells, either in gel emulsion with beads or in wells, then quantitatively converting RNA molecules by adding a unique molecular identifier (UMI). Since there are no known barcodes for droplet-based single-cell technologies like Drop-seq, a safe list of variants ensures interoperability. A well-based technique called ICELL8 allows for the selection of subsets of wells by known barcodes7 and has a known barcode whitelist. Dual indexing and full-length RNA sequencing are other well-established technologies used in SmartSeq3. These represent a variety of technological classes with various cell barcode processing setups, including Chromium. UniverSC and the pipeline used in the initial publication of the approach were both run on datasets of human cell lines in order to gauge the degree of similarity between UniverSC and the pipelines for these four technologies (Chromium, Drop-seq, ICELL8, and SmartSeq3). similarity
The mentioned pipelines were specifically compared to UniverSC:
- Cell Ranger for Chromium data,
- dropSeqPipe for Drop-seq data,
- CogentAP for ICELL8 data, and
- zUMIs for SmartSeq3 data.
When it comes to data integration, using UniverSC on all datasets from various platforms is preferable to using separate pipelines for each technology. Given the great degree of connection between the outputs of UniverSC and the different other pipelines examined, as well as the fact that all pipelines operate within a similar framework, a significant influence cannot be anticipated. However, applying UniverSC to all samples results in quantifiable gains in data integration when compared to using separate processes on datasets produced by various platforms.
As single-cell techniques become an integral part of various studies, it becomes necessary to mitigate technical errors and integrate scRNAseq data generated by different groups and platforms. It is convenient and essential to process data containing various barcodes and UMI configurations in a consistent framework. Although there are pipelines that can be configured for different technologies (dropSeqPipe, zUMI, dropEst, Kallisto/BUStools), Cell Ranger works well in server or cluster environments and produces rich and informative output summaries. Note that UniverSC uses Cell Ranger version 3.0.2 for licensing reasons.
A new version of Cell Ranger is now available, but these updates do not significantly impact scRNAseq data processing, as changes in the core allow analysis beyond scRNAseq. B. scATAC-seq, TCR, and BCR analysis. As new single-cell techniques are developed, the utility of UniverSC eliminates the need to develop proprietary technology-specific data processing pipelines.
Compliance of UniverSC with Cell Ranger
UniverSC requires a similar set of input parameters as Cell Ranger, in addition to two inclusions.
- Paired-end FASTQ input files and
- Cell Ranger-prepared reference data
As is typical in 3′ scRNAseq methods, UniverSC, by default assumes that Read 1 of the FASTQ contains the cell barcode, UMI, and Read 2 has the transcript sequences that will be mapped to the reference. Given a known barcode and UMI length, UniverSC will examine the file name and barcodes, making any necessary adjustments to the settings to make them compatible with Chromium.
The containerized graphical application, docker image, and command-line tool provide consistent and thorough integration, comparison, and assessment across data generated from a multitude of platforms.
Strengths of UniverSC
The UniverSC documentation is available as a manual aid tool in the terminal and user interface that verifies file inputs and displays error messages to indicate potential issues. Both the source code and a docker image for UniverSC are made accessible to the public, and it may be used on any Unix-based machine in the shell. The user can also choose to install a GUI for UniverSC.
Curbing technical mistakes and integrating scRNAseq data collected across multiple organizations and platforms will be required as single-cell technologies become essential to a wide range of research. It will be practical and necessary to process data with different barcode and UMI configurations inside a unified framework.
The usefulness of UniverSC eliminates the requirement to create a specific data processing pipeline for their own technology when novel single-cell technologies are created.
Last but not least, it will allow for a fair comparison when assessing the best platform for a particular sample type, which may be crucial with difficult samples such as those that include giant cells or digestive enzymes.
Final thoughts on UniverSC
A unifying tool is required to integrate several scattered publicly accessible datasets by processing the data according to the same processes and settings. This is due to the growing number of scRNAseq datasets produced using various platforms and published by laboratories internationally. Documentation and codes used to generate each filtered/downsized dataset are provided in the UniverSC GitHub repository. The complete raw output datasets are available for Chromium, Drop-seq, and ICELL8.
Article Sources: Reference paper
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Riya Vishwakarma is a consulting content writing intern at CBIRT. Currently, she's pursuing a Master's in Biotechnology from Govt. VYT PG Autonomous College, Chhattisgarh. With a steep inclination towards research, she is techno-savvy with a sound interest in content writing and digital handling. She has dedicated three years as a writer and gained experience in literary writing as well as counting many such years ahead.