The World Health Organization (WHO) appraises that 700,000 individuals now die yearly because of antibiotic resistance and anticipates that this number will surpass 10 million every year by 2051.
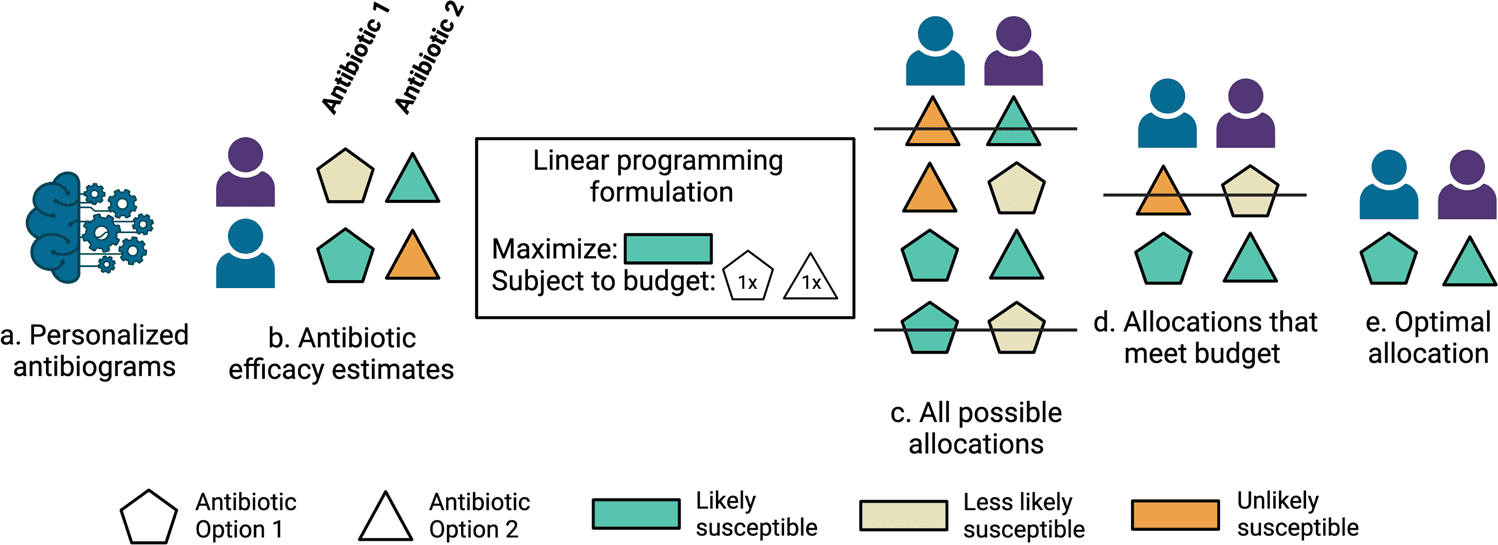
Stanford University scientists developed machine learning models that predict antibiotic susceptibility patterns (personalized antibiograms) using electronic health record data. Personalized empiric antibiotic subscription with personalized antibiograms may enhance patient safety and antibiotic stewardship by decreasing the unnecessary use of broad-spectrum antibiotics that breed a growing tide of resistant organisms.
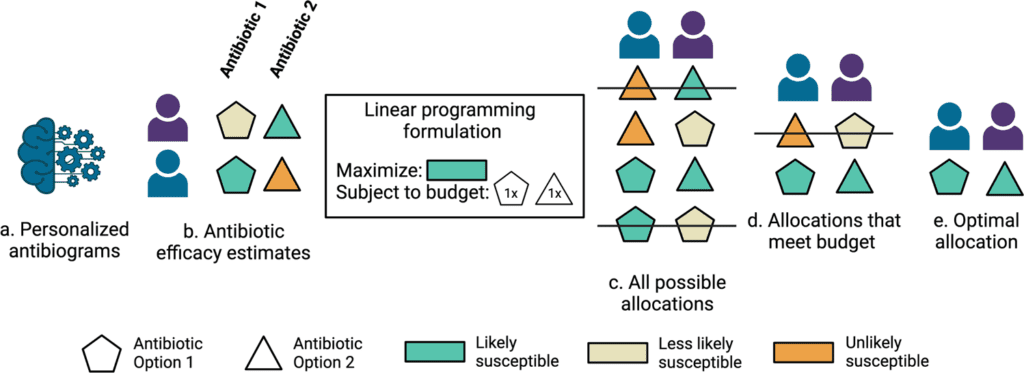
Image Source: Personalized antibiograms for machine learning-driven antibiotic selection.
Increasing antibiotic resistance is a normal and inevitable result of customary antibiotic use, raising the approaching danger of a post-antibiotic era that could cripple routine clinical consideration with higher infection-related mortality and expenses of care.
Recognizing Advanced Antibiotic Prescribing
The Centers for Disease Control and Prevention (CDC) recognizes further developing antibiotic prescribing through antibiotic stewardship as the main activity to battle the spread of antibiotic-resistant bacteria.
For instance, 60% of the hospitalized patients get antibiotics, notwithstanding that half a portion of antibiotic medicines are of little to no importance, meaning that the antibiotic use was unwarranted, some wrong antibiotic was administered, or the antibiotic was administered with the incorrect portion or duration.
A key challenge is that antibiotic agents should frequently be prescribed empirically before the identity of the infective organism and antibiotic susceptibilities are known. Microbial cultures are the definitive diagnostic tests for this data, yet may take days to affirm results, extremely lengthy to defer initial therapy.
Broad-Spectrum Antibiotics
Broad-spectrum antibiotics assist with guaranteeing the inclusion of a range of organisms that would prompt rapid clinical deterioration if left untreated. However, it is definitively the excessive utilization of antibiotics that increments antibiotic-resistant microorganisms.
Overuse of broad-spectrum antibiotic agents can consequently have serious instant and indirect outcomes, including expanding antibiotic resistance from drug-specific toxicities and secondary infections like Clostridioides difficile colitis.
Choosing Empiric Antibiotics for Existing Standards of Care
Existing principles of care for choosing empiric antibiotic agents include alluding to clinical practice rules along with information on the institution-explicit antibiograms-a yearly report from an institution’s microbiology laboratory that tracks the most well-known organisms isolated by microbial cultures and the percentages that were susceptible to various antibiotics.
An institution’s antibiogram could report, for instance, that 1000 Escherichia coli were isolated in the earlier year and that 98% were susceptible to meropenem, while just 89% were susceptible to ceftriaxone. These methodologies may not include several or any persistent patient-specific highlights. Microbial culture results found inside the electronic health record can be utilized to objectively quantify whether picked antibiotic agents were appropriate, however assuming that options would be sufficient.
Here, the scientists hypothesized that the care routine might take advantage of machine learning-based clinical decision support for customized treatment suggestions.
Computational Clinical Decision Support
The advancement of computational clinical decision support for antibiotic prescribing stems back a very long time to any semblance of MYCIN and Evans et al.- rule-based frameworks that guide clinicians through empiric antibiotic selection.
Though promising, neither one of the frameworks was broadly taken on by clinicians as they were not effortlessly incorporated into their medical workflow or adjustable to continually advancing local antibiotic resistance patterns.
With advanced clinic IT and electronic medical record software, it is presently conceivable to coordinate clinical decision support into medical work processes and dynamically train models with real-world clinical data streams.
Literature concerning cutting-edge data-driven ways to deal with antibiotic decision support falls into two particular classifications. One classification of studies predicts infection status when microbial cultures are ordered, offering promising consideration for when antibiotics are required.
Limitations in the vast majority of these earlier studies are that positive microbial culture results were utilized as a proxy for the outcome of infection, notwithstanding their being both false positive and false negative microbial cultures regarding an actual clinical infection. In addition, these examinations don’t resolve the subject of which antibiotics ought to have been administered.
The second classification of studies predicts antibiotic susceptibility results for positive microbial cultures. These investigations address the test of choosing the right antibiotic. Antibiotic prescribing policies that influence machine learning predictions were simulated and benchmarked against retrospective clinician prescribing and proposed better performance.
Optimizing patient coverage rates, in any case, is just a single significant objective that could be naively addressed by prescribing maximally broad antibiotics to all patients without consideration for harmful consequences for the individual or populace. A further imperative study needs to efficiently assess the trade-off between boosting antibiotic coverage across a populace of patients and limiting broad-spectrum antibiotic use.
Personalized Antibiograms
The researchers trained binary machine learning models (also known as personalized antibiograms) utilizing structured electronic health record information to gauge the probability that infection would be susceptible to 12 common empiric antibiotic decisions (four of which were combination treatments or combination therapies).
Labels were obtained from the antibiotic susceptibility reports of the microbial cultures. Observations by the scientists were then allocated a positive label when organisms isolated were considered susceptible (not intermediate or resistant) to the respectively tested antibiotics in light of microbiology laboratory principles.
Promotion of Antibiotic Stewardship with the help of Reduced Broad Spectrum Antibiotic Usage
To assess the potential for decreasing broad-spectrum antibiotic agents with personalized antibiograms, the scientists sorted antibiotic selection as per their antibiogram value (fraction of infections recorded as susceptible) and rehash the linear programming based optimization simulations under revised constraints where the personalized antibiogram based treatment selection is compelled to utilize less broad-spectrum (higher antibiogram value) antibiotics instead of narrower range (lower antibiogram value) antibiotics.
The scientists tracked the coverage rate achieved by a personalized antibiogram and matched it to the coverage rate achieved by the clinician and random selections.
Analysis of Sensitivity
The researchers directed a sensitivity analysis with the Stanford cohort to incorporate patients with negative microbial culture results into their model performance estimates.
This was finished by
- Building an electronic phenotype to flag patients with negative microbial cultures that lacked infection,
- Training twelve new personalized antibiogram models (sensitivity analysis models) that incorporate patients with negative cultures, and
- Utilizing inverse probability weighting model performance estimates
The Demonstration of Antibiotic Susceptibility Classifiers
The antibiotic susceptibility classifiers (personalized antibiograms) showed modest to moderate discriminatory power concerning AUROC, consistent with prior state-of-the-art literature.
However, AUROC alone is not a decent determinant of clinical utility. The scientists’ optimization simulations exhibit that even with modest AUROCs, antibiotic selection informed by personalized antibiograms can coordinate or surpass clinician execution.
Besides, antibiotic selection directed by personalized antibiograms accomplished comparable coverage rates to those found in reality with less broad-spectrum antibiotics, a continuous and essential antibiotic stewardship challenge.
The Endpoint
The machine learning classifiers trained by using electronic health record information can predict antibiotic susceptibility for patients with positive microbial cultures.
Antibiotic selection strategies directed by personalized antibiograms could keep up with or further develop infection coverage rates while utilizing less broad-spectrum antibiotic agents than are found in real-world practice.
Machine learning-driven antibiotic selection could work on antibiotic stewardship without forfeiting and possibly, in any event, improving patient health.
Finally, not all infections present a similar threat to patients whenever treated with an improper antibiotic routine. Future work might take advantage of utilizing the possibility that a few infections are more complex to treat than others.
Paper Source: Corbin, C.K., Sung, L., Chattopadhyay, A. et al. Personalized antibiograms for machine learning driven antibiotic selection. Commun Med 2, 38 (2022). https://doi.org/10.1038/s43856-022-00094-8
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Tanveen Kaur is a consulting intern at CBIRT, currently, she's pursuing post-graduation in Biotechnology from Shoolini University, Himachal Pradesh. Her interests primarily lay in researching the new advancements in the world of biotechnology and bioinformatics, having a dream of being one of the best researchers.