The investigation of nuclear anomalies and how they influence the regulation of genes warrants analysis of various cellular functions and diseases. The researchers from MIT, Harvard, and the University of California San Francisco describe a new approach, Expansion in situ Genome Sequencing (ExIGS), that allows, for the first time, to unravel the relationship between nuclear aberrations and spatial organization of chromatin inside the nucleus. ExIGS integrates expansion microscopy or ExM and in situ genome sequencing or ISGS, which allows for the simultaneous imaging of nuclear proteins and genomic DNA in a high-resolution manner. This amazing technology will allow scientists to routinely analyze how alterations in nuclear shape affect chromatin arrangement and gene expression regulation.
Grasping Nuclear Defects
Nuclear abnormalities, also referred to as nuclear alterations, consist of changes in the size and shape of the nucleus and alteration of the nuclear chromatin’s topological arrangements. This generally occurs during disease conditions such as cancers and various hematologic disorders. These abnormalities may interfere with the normal genome organization as well as function, thereby altering gene expression, which may cause malfunctioning of the cells.
The Role of Spatial Chromatin Organization
Chromatin’s three-dimensional positioning inside the cell nucleus is essential in enhancing gene expression, providing genome stability, and regulating the cell cycle. Active transcription of chromatins shows localization to transcriptional hubs, while inactive transcriptional chromatins usually occur at the nuclear lamina. Changes in this exact organization have been associated with aging and disease-observable characteristics.
ExIGS: A Revolutionary Technique
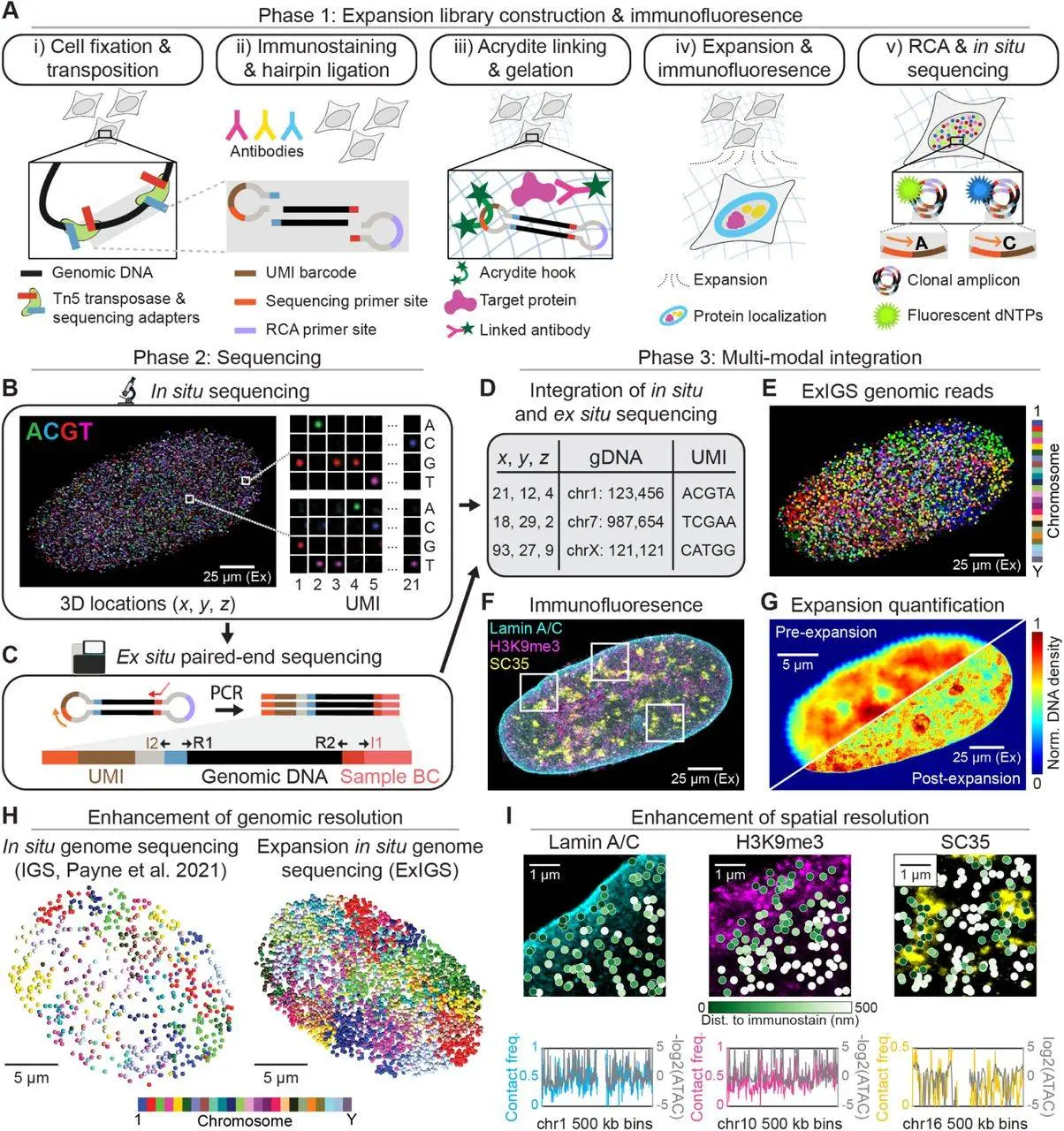
ExIGS (Expansion in situ Genome Sequencing) is an advanced method that fuses expansion microscopy and in situ genome sequencing very well. With this new technique, it is possible to visualize within single living cells and, at super-resolution, the organization of genomic DNA and nuclear proteins.
Expansion Microscopy
- Principle: This is a cellular inclusion technique whereby cells or tissues are embedded in a gel-like tissue, and a gel is stretched, hence the name expansion microscopy. Consequently, this expansion maintains the relative positions of entities of interest within the specimen.
- Benefits: By expanding the sample, ExIGS allows improved image size and exposure of the specimen’s minute components, which are otherwise impossible, which is made possible.
In Situ Genome Sequencing
- Principle: This is a simple procedure of determining the sequence of genomic material without removing it from the cell. By doing so, the spatial arrangement of the molecules can be maintained.
- Benefits: By sequencing DNA in situ, researchers can study the spatial distribution of genes and regulatory elements within the nucleus.
ExIGS Workflow
The ExIGS workflow includes the following:
- Sample Preparation: Tissue or cell samples are treated and packaged in a gelatinous compound.
- Expansion: The gel is expanded to increase its size.
- In Situ Genome Sequencing: The process of directly sequencing genomic DNA from the expanded cells.
- Immunofluorescence: Nuclear proteins are labeled and imaged with fluorescent antibodies.
- Data Integration: The sequence and imaging data are used to examine the spatial relationships between genomic regions and proteins.
Key Findings from the Study
The main goal of the researchers was to apply ExIGS on progeria fibroblasts, a cell type associated with nuclear abnormalities. They found out:
Lamin Abnormalities: There are abnormal lamin proteins and nuclear inner membrane proteins in progeria fibroblasts. These changes are accompanied by changes in the shape and size of the nucleus.
Disrupted Euchromatin Organization: Lamin abnormalities lead to disruptions in the spatial organization of euchromatin, the transcriptionally active form of chromatin.
Repressive Environment: Abnormality of Lamin causes an unusual nuclear environment that is more repressive, and gene activity is lowered.
Tissue Variation: Other tissues tend to have the same lamin abnormalities and effects, suggesting that there is a general tendency in this behavior.
Conclusion
Expansion in situ genome sequencing provides exciting prospects in understanding how nuclear abnormalities translate spatial organization of the chromatin. This technique reveals new avenues into the pathogenesis of different diseases at a molecular level and offers great potential for the future. The development trajectory of ExIGS will likely make it relevant in the growth of knowledge on nuclear structure and the progression of diseases.
FAQs
In situ sequencing refers to a method that enables the sequencing of nucleic acids, i.e., DNA or RNA, without extracting them out of living organisms or biological tissues. In contrast to conventional sequencing techniques, which involve the separation of nucleic acids from cells, in situ sequencing allows for keeping the spatial information of the molecules, which is preventive and constructive.
In situ genome sequencing (IGS) is a novel method that enables the simultaneous sequencing and imaging of genomes within intact biological samples, allowing researchers to explore the spatial organization of genetic material at high resolution. This technique integrates DNA sequencing with three-dimensional spatial context, addressing limitations of existing methods that either lack base pair resolution or direct spatial localization. By applying IGS to samples like human fibroblasts and early mouse embryos, scientists can spatially localize thousands of genomic loci, characterize changes in genome structure, and reveal chromatin domains across different developmental stages. The workflow involves creating in situ sequencing libraries, multimodal sequencing, and computational integration of genetic and spatial information, ultimately bridging the gap between genomic sequence and structural organization within cells.
ExIGS has a wide range of potential applications, including:
– Studying the spatial organization of chromatin
– Analyzing nuclear abnormalities
– Investigating disease mechanisms
– Developing new therapeutic strategies
Article Source: Reference Paper |All code available on GitHub.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.