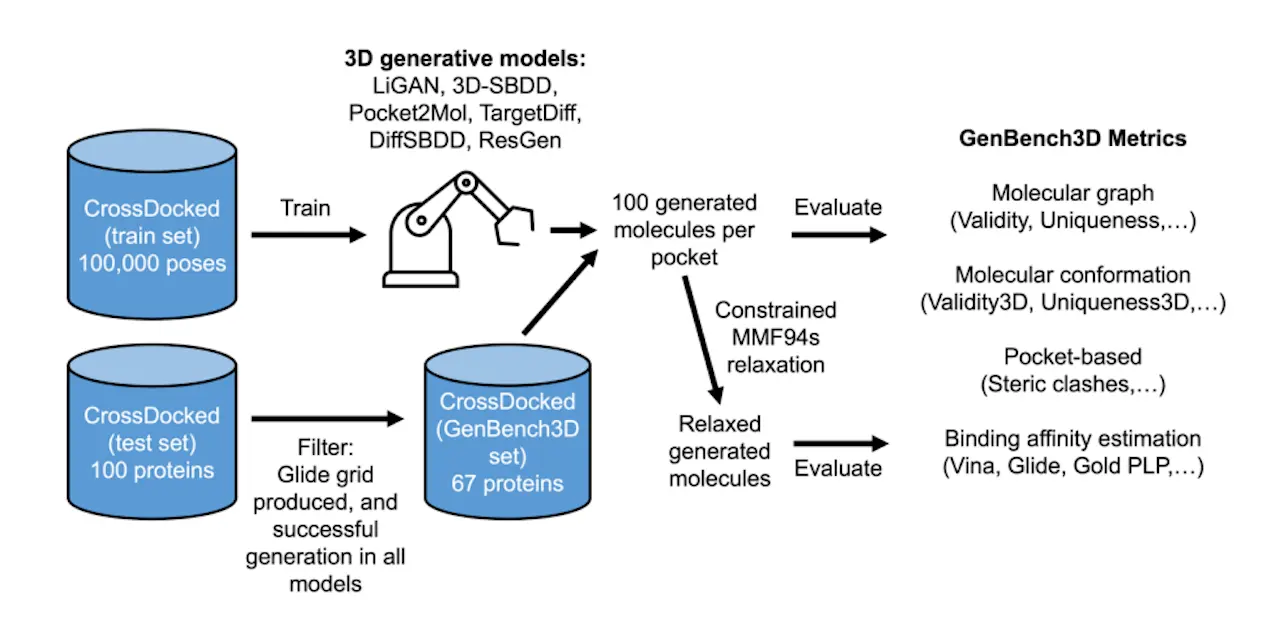
Evaluating specific properties of molecules from simple SMILE strings isn’t enough. Instead of simple chalkboard formulae, our aim should be to conjure up three-dimensional molecular structures that could fit snugly into tiny pockets within proteins. 3D deep molecular generative models stand out by enabling the generation of molecules based on 3D-dependent properties, such as binding affinity within specific protein binding pockets. These models present a significant advancement over traditional generative methods that rely on SMILES or molecular graphs, which lack the capacity to assess the quality of the generated molecular conformations directly. Recognizing this gap, Baillif et al. from the University of Cambridge developed GenBench3D, a novel benchmark designed to evaluate the performance of 3D generative models in producing valid molecular conformations.
Why do we need GenBench3D?
There is a requirement for deep molecular generative models, where molecules are crafted not just for their chemical properties but also for how well they can interact with the specific binding pockets. Traditional tools for evaluating molecular models, like GuacaMol or MOSES, have been fantastic at assessing 2D molecular generators. However, these benchmarks fail to determine the 3D quality of generated molecules, a critical aspect of structure-based drug design.
GenBench3D addresses this by introducing the Validity3D metric, which evaluates the conformation quality based on the likelihood of bond lengths and valence angles derived from the Cambridge Structural Database (CSD). This approach ensures generated molecules adhere to realistic geometric constraints observed in empirical data.
Methodology and Metrics
Validating Molecular Graphs
Firstly, a molecule must be deemed valid according to RDKit, a prominent cheminformatics software suite. A molecule is considered to have a valid graph if it can be successfully parsed by RDKit v.2023.6 with default sanitization. This sanitization process involves standardizing non-standard valence states, Kekulizing aromatic rings, assigning radical electrons, setting aromaticity, conjugation, and hybridization, and verifying valence correspondence. To quantify this, GenBench3D calculates the molecular graph validity (V_graph).
Ensuring Uniqueness
Next, it is essential to verify that the model isn’t simply producing the same molecule repeatedly. Molecular graph uniqueness (U_graph(N)) is measured as the fraction of unique molecular graphs among the first N-generated molecules with a valid graph.
Assessing Novelty
Innovation is vital, and GenBench3D evaluates whether the models generate new molecules that were not seen during training. Molecular graph novelty (N_graph) is measured as the fraction of molecules with valid graphs absent from the training set.
Benchmarking Generative Models
GenBench3D benchmarks six structure-based 3D generative models: LiGAN, 3D-SBDD, Pocket2Mol, TargetDiff, DiffSBDD, and ResGen. These models employ various architectures, including variational auto-encoders (VAEs), graph neural networks (GNNs), and diffusion models, to generate molecules within binding pockets.
The critical metric introduced by GenBench3D, Validity3D, assesses the geometric validity of generated molecular conformations. This metric relies on the q-value, a ratio of the likelihood of a query value to the maximum likelihood based on CSD data. The s-value, a geometric mean of these q-values, provides a single score for the overall geometric validity of a conformation. A molecule is deemed 3D-valid if it exhibits likely bond lengths and valence angles and lacks intramolecular steric clashes.
Results
The benchmarking results revealed that up to 11% of the generated molecules across the evaluated models had valid conformations. This highlights a significant challenge in the current state of 3D molecular generative models. However, the study demonstrated that performing local relaxation of the generated molecules within the binding pocket improved the Validity3D scores significantly, with a minimum increase of 40% across all models.
Models like LiGAN, 3D-SBDD, and TargetDiff showed higher (worse) Vina scores for valid relaxed molecules compared to raw ones. This indicates that initially generated conformations might fit into the pocket more snugly but at the cost of unrealistic geometric distortions. In contrast, Glide scores, which place higher importance on ligand strain, showed better results for valid relaxed molecules, underscoring the importance of considering ligand strain in affinity predictions.
Among the models, TargetDiff and Pocket2Mol performed better in terms of median Vina, Glide, and Gold PLP scores after local relaxation, suggesting their superior ability to generate geometrically valid and biologically relevant conformations.
Conclusion
GenBench3D offers a comprehensive and robust benchmark for evaluating the performance of structure-based 3D molecular generative models. The introduction of the Validity3D metric provides a critical tool for assessing the geometric validity of generated molecular conformations, addressing a significant gap in traditional benchmarks. The findings of this study reveal the current challenges in 3D molecular generation and highlight the importance of continuous improvement in model development and evaluation. As computational methods continue to advance, benchmarks like GenBench3D will play an important role in guiding the development of more accurate and effective generative models, ultimately accelerating the discovery of new drugs and therapeutics.
Article Source: Reference Paper | The suite of metrics used in this benchmark is implemented in the GenBench3D Python package, available on GitHub.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Neermita Bhattacharya is a consulting Scientific Content Writing Intern at CBIRT. She is pursuing B.Tech in computer science from IIT Jodhpur. She has a niche interest in the amalgamation of biological concepts and computer science and wishes to pursue higher studies in related fields. She has quite a bunch of hobbies- swimming, dancing ballet, playing the violin, guitar, ukulele, singing, drawing and painting, reading novels, playing indie videogames and writing short stories. She is excited to delve deeper into the fields of bioinformatics, genetics and computational biology and possibly help the world through research!