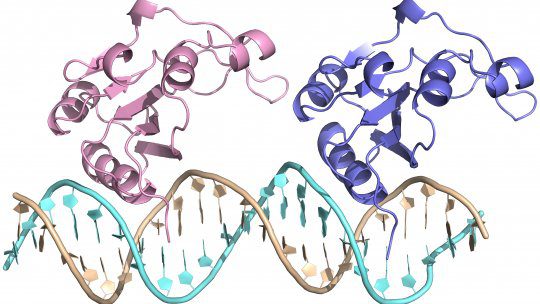
Scientists from Barcelona have discovered a new hexameric protein structure for the RepB protein, which is involved in catalyzing replication initiation in Streptococcal plasmid pMV158, conferring antibiotic resistance to tetracycline. The new hexameric structure, obtained using biochemical methods and X-ray crystallography, is found to render flexibility, which attributes to the dual functionality of the protein, viz., binding to two distinct sites of the plasmid and separating one of the DNA strands by cutting it off, resulting in the initiation of DNA replication. The new structural information now available will aid in designing new antibiotics for therapeutics as well as in a better understanding of antibiotic resistance in the laboratory.
The story of antibiotic resistance and DNA replication featuring RepB protein
Antibiotic resistance results when the disease-causing bacterium or other pathogens acquire genetic mutations to adapt against the drugs designed to nullify them. These mutations, under selective pressure to adapt to the toxic environment, as generated by the drugs, result in drug-resistant variants. These acquired mutations are often not vertically obtained from previous generations but rather are acquired horizontally through horizontal gene transfer (HGT). This lateral transfer of mutations is across species in the tree of life and is quite prevalent in prokaryotes like bacteria and some eukaryotes like yeast. Antibiotic resistance genes in bacteria are mostly acquired by HGT.
The mechanism of HGT in bacteria typically involves plasmids. Plasmids are circular pieces of DNA that are separate from the chromosomal DNA and are capable of independent replication.DNA replication, as we know, takes place in all living organisms and is the basis for genetic inheritance. Plasmids act as scaffolds for antibiotic-resistant genes, which are either transferred over generations (vertically) or across species (horizontally).
Plasmids replicate independently using the DNA replication toolbox, which includes the replication initiation protein, Rep protein. Rolling circle replication (RCR) is one of the mechanisms by which plasmids replicate. The replication initiation protein Rep interacts with the specific DNA sequences within the plasmid known as the double-strand origin (dso) for the initiation of rolling circle replication (RCR). The bind locus is where the Rep protein binds to and severs a specific bond within the nick sequence. The now-bound Rep protein thus initiates leading strand replication. RepB is the replication initiation protein of the Streptococcus plasmid pMV158.
The crystal structure of RepB has been previously determined. It is purified as a homohexamer. Individual monomers have an oligomerization domain (OD) and an origin binding domain (OBD), which are connected via a short hinge region. The hexameric structure of the protein is due to the OD. The OBD houses the catalytic site for recognizing, cleaving, and joining dso DNA. The bind locus comprises three tandem direct repeats known as distal direct repeats (DDR). RepB binds primarily to DDRs. The scientists, led by Dr. Miquel Coll at the IRB Barcelona, present a new hexameric structure for the RepB protein, which shows a completely different organization of the OBDs as compared to previously determined structures but identical 6-fold symmetry within the ODs.
Experiments to determine the new structure
The authors implemented several biochemical techniques coupled with X-ray crystallography to arrive at the novel protein structure for RepB. The several steps are briefly described:
- To determine the cooperative binding of the monomeric OBD to the entire bind locus, Electrophoresis mobility shift assays (EMSA) were performed.
- Other biochemical methods involved Nicking and strand transfer experiments, footprinting experiments, and Surface plasmon resonance (SPR)experiments.
- Full-length RepB purified crystals were obtained by implementing a hanging-drop vapor-diffusion method.
- Crystallization of the OBD was also obtained using a hanging-drop vapor-diffusion method.
- Crystallization of the OBD-DNA complex was obtained using a sitting-drop vapor-diffusion method.
The outcome of the study
- New RepB hexamer crystal structure: The authors have solved a new structure of full-length RepB with structural versatility resulting in dual functionality of the protein. While the OD remains structurally identical to previously determined structures of RepB, the OBDs show high variability in structural conformations.
- OBD binding to the entire bind locus does not show cooperative behavior, which explains the lack of protein-protein interactions between monomers of the OBD-DDR complex.
- RepB structure is shown to exhibit two different DNA interaction sites within the OBD, namely the recognition helix area and the active site area.
Conclusion
The authors have solved a novel structure for the RepB protein involved in RCR initiation. The new hexamer structure was found to render flexibility to the protein, resulting in its dual functionality of binding to two different sites of the plasmid and cleaving off one of the strands of DNA, thus initiating DNA replication. This is indeed a remarkable discovery in terms of understanding antibiotic resistance and designing novel drugs for diseases caused by bacteria and other pathogens. While the human population is posed with a global threat of global antibiotic resistance, which could lead to severe novel infections arising often, this new discovery can be a sigh of relief towards better dealing with antibiotic-resistant strains and a leap towards better health for all of humanity.
Article Source: Reference Paper | Reference Article
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Banhita is a consulting scientific writing intern at CBIRT. She's a mathematician turned bioinformatician. She has gained valuable experience in this field of bioinformatics while working at esteemed institutions like KTH, Sweden, and NCBS, Bangalore. Banhita holds a Master's degree in Mathematics from the prestigious IIT Madras, as well as the University of Western Ontario in Canada. She's is deeply passionate about scientific writing, making her an invaluable asset to any research team.