Antibiotic resistance is increasing as a global problem, meaning previously effective treatments for bacterial infections have ceased to work. It is vital to accurately identify the antibiotic resistance profile of an infection to choose the right antibiotic and improve patient outcomes. In Germany, researchers from the Technical University of Munich (TUM) are exploring this possibility using real-time genomics.
Antibiotic-resistant gene detection by conventional means can be slow and may not even reveal hidden resistance genes at times. This delay could result in inappropriate treatment or the patient’s worsening condition. The TUM study aimed at identifying hidden antibiotic resistance genes in a patient with complex bacterial infection by real-time genomics with nanopore sequencing.
The Case: Discovered Resistance Gene
Fever was the first symptom that led to the diagnosis of carbapenem-resistant Klebsiella pneumoniae in the patient involved in the study. They are a kind of wide-spectrum antibiotics that can be used as a last resort treatment for complicated bacterial infections. The clinical team began initial therapy with Meropenem, which is a carbapenem antibiotic. As a result, this case worsened upon changing to Ceftazidime-Avibactam (CAZ-AVI), another antibiotic typical to carbapenem-resistant K. pneumoniae.
For this reason, all other usual diagnostic approaches available at the clinic, such as VITEK2 for testing antibiotic resistance and MALDI-TOF MS for identifying bacteria, did not explain why this patient was getting sicker and sicker. However, real-time genomics through nanopore sequencing revealed a hidden cause – a low-abundance plasmid harboring the blaKPC-14 gene variant. These variations specifically lead to CAZ-AVI resistance but may recover sensitivity towards Meropenem-like carbapenems, thus giving reasons for the patient’s healing response.
Faster and Better Real-Time Genomics?
In comparison to traditional methods, the study points out that real-time genomics has tangible merits. Here is a list of various distinctions:
Speed: Real-time genomics can give answers within hours, which is much faster than the days spent in traditional ways. This prompt turnaround enables doctors to make quick changes in treatment plans that could consequently assist patients in becoming better.
Accuracy: The research posits that real-time genomics may be more accurate in detecting hidden resistance genes, especially those found on low-abundance plasmids. These genes can go unnoticed through traditional approaches.
Adaptive Sequencing: Real-time genomics allows for “adaptive sequencing.” Samples may be sequenced as long as needed until reliable diagnosis is achieved. In complex infections, this becomes particularly useful as traditional methods might not yield sufficient information.
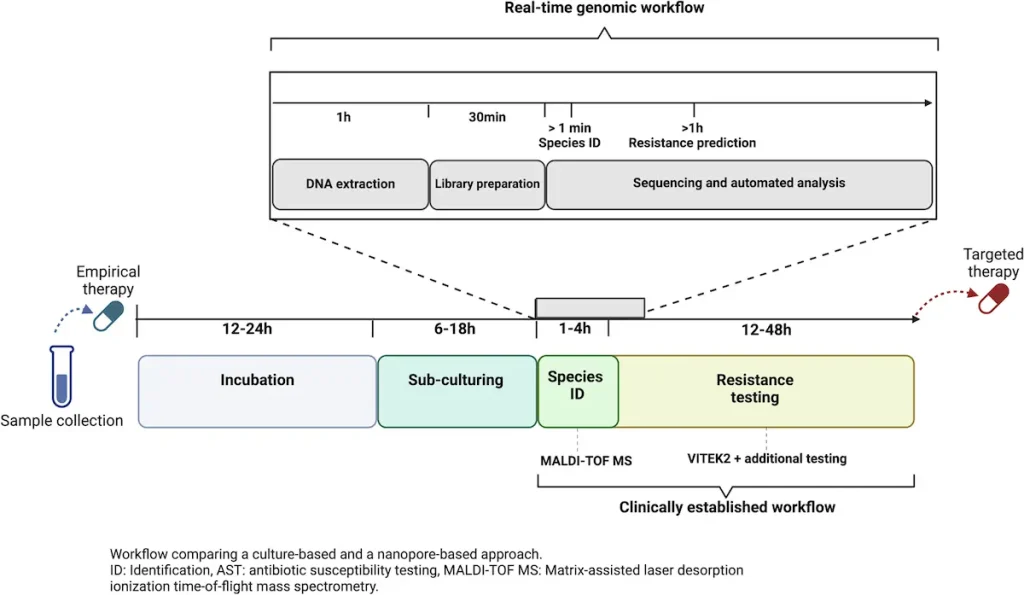
Boundaries and What Comes Next
In this context, while the study demonstrates considerable promise for real-time genomics, it has limitations.
The Emphasis on Isolates: The Research’s main concentration was bacterial isolates, thereby overlooking all the other microorganisms in an infection. On the other hand, metagenomic sequencing that interrogates an entire genetic material from a sample would have given a comprehensive result.
The Insufficiency of Data: It is also important to note that this study was retrospective and involved only one patient. For its applicability to be generalized, larger prospective studies should be done.
However, despite these drawbacks, the study provides a fascinating look into what might happen in the field of antibiotic resistance diagnostics in the future. More research will be needed to link real-time genomics with standard methods and evaluate its performance across different clinical settings. Nanopore sequence technology’s cost-effectiveness and portability are particularly advantageous when access to sophisticated diagnostics is restricted by limited financial resources in low-income countries.
Beyond real-time genomics: the dynamic landscape of antibiotic resistance diagnostics
The study from TUM on potential real-time genomics with nanopore sequencing is a big step forward. Nonetheless, tackling antibiotic resistance requires a multifaceted approach. Here are some emergent trends that will determine the future of diagnostics in this field:
Artificial Intelligence (AI) in Diagnostics:
Machine learning algorithms have been trained with large genomic and clinical datasets, allowing AI to identify patterns and predict antibiotic resistance profiles with higher precision. AI could potentially analyze complex data from real-time genomics and traditional methods to create a more holistic picture of an infection.
Multiplex PCR Panels:
These quick diagnostic tests can simultaneously check for several resistance genes. Even though they are not as exhaustive as real-time genomics, they are faster and more targeted than traditional approaches. Particularly, multiplex PCR panels could be useful in low-resource settings or where there is a need for quick turnaround time for results.
Point-of-Care Diagnostics:
To begin with, developing portable diagnostic tools that are easy to operate and yield results at the point of care is very important for early and effective treatment, especially in remote areas. These devices can detect antibiotic resistance markers directly from clinical samples in various ways, including using microfluidics and biosensors.
Metagenomic Sequencing:
Metagenomic sequencing, on the other hand, offers a more comprehensive view of what is happening within an infection, as highlighted by the TUM study limitation; it is not only cultured bacteria but also potential pathogens like fungi, viruses, and others. In addition, metagenomic sequencing examines every gene present in a sample, thus giving a chance to find hidden resistance mechanisms and co-infections that conventional methods might miss.
Antimicrobial Stewardship Programs:
Resistance trends are monitored based on evidence-based prescribing guidelines that promote the judicious use of antibiotics through these programs. Real-time genomic data would, therefore, get integrated into such schemes to enable the proper selection of antibiotics while limiting the emergence of new resistance mechanisms.
Conclusion
Combating antibiotic resistance requires a multi-pronged approach. A promising one is real-time genomics using nanopore sequencing, although just one element in the jigsaw. We can develop more inclusive and efficient diagnostic landscapes by combining this technology with artificial intelligence, multiplex PCR panels, point-of-care diagnostics, and robust antimicrobial stewardship programs. This joint effort through continuous research and development will stop the spread of antibiotic resistance, thus ensuring that lifesaving drugs continue to work.
Join the Discussion
Antibiotic resistance is becoming a danger to public health. This blog post aimed at revealing how real-time genomics, among other emerging diagnostic tools, could help us fight against resistant bacteria.
Do you know anyone who has suffered an antibiotic-resistant infection?
What are your worries regarding these forms of drug resistance? Real-time genomics sounds expensive. How do we ensure access to this technology to all population groups?
We’d love to hear your thoughts! Share your comments below.
Article Source: Reference Paper | All computational scripts are available on GitHub.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.