A major component of the expanding significance of precise nucleic acid quantification in molecular biology is its emphasis on the field’s application in genomic research and infectious disease diagnosis. This review looks at recent developments in digital loop-mediated isothermal amplification (dLAMP) and digital polymerase chain reaction (dPCR), which provide improved sensitivity and absolute quantification, respectively, and overcome the drawbacks of conventional Nucleic Acid Amplification Testing (NAAT). The evaluation provides an overview of the significant progress made by dNNAT in tackling urgent public health concerns, particularly in light of the COVID-19 epidemic. Researchers also look at how artificial intelligence (AI) is revolutionizing dNAAT image analysis, which increases accuracy and efficiency and overcomes conventional limitations about cost, complexity, and data interpretation.
Introduction
Nucleic acids, the fundamental arbiters of genetic information, have been crucial in scientific endeavors since the inception of life around 3.5 billion years ago. The COVID-19 pandemic, which has impacted over 711 million people globally, has highlighted the need for rapid and precise quantification of nucleic acids. Ultrasensitive detection methods, such as Nucleic Acid Amplification Tests (NAAT), are essential for preventing infectious diseases like malaria, dengue, tuberculosis, and monkeypox. As little as ten copies of the target can be detected per response by NAATs, which are incredibly sensitive and specific. Digital Polymerase Chain Reaction (dPCR) and digital Loop-mediated Isothermal Amplification (dLAMP) are two recent innovations that have revolutionized nucleic acid quantification. These innovations have led to a shift towards absolute quantification, which can detect precise gene expression levels down to a single molecule and improve the detection of viral particles. This is true for genetic disorders, cancer, and virology.
dPCR vs. Conventional PCR
Conventional PCR amplifies DNA in a single reaction chamber, whereas dPCR splits the DNA sample into millions or thousands of separate, concurrent reactions in microreactors. Target DNA molecules could be present in these processes, or they might not be. Each partition is inspected upon amplification to see whether the amplification product identified by fluorescence is present or absent. Poisson statistics are employed in conjunction with the fraction of positive reactions to ascertain the precise quantity of target molecules present in the initial sample. Amplification bias is subverted by the absolute quantification of dPCR, which increases reliability over traditional PCR and quantitative PCR (qPCR). dPCR can distinguish between a 1% variation in chromosome copy number and identify fewer than one copy per 100,000 wild-type sequences with up to 1,000,000 reactors and a dynamic range of 107.
Understanding Nucleic Acid Amplification Testing (dNAAT)
dNAAT is a breakthrough in molecular biology that allows nucleic acid quantification to be done in absolute terms by dividing samples into multiple unique units. This makes it possible to count target sequences directly. For accurate amplification, dNAAT partition samples using thousands of compartments were created using droplet-based, chip/chamber-based, and agarose-based approaches. A systematic procedure of sample dilution, partitioning, amplification, and in-depth analysis is followed by these techniques. When it comes to producing droplets the size of picolitres, meeting high-throughput demands in microwell dPCR, reducing the chance of contamination, and preserving sample stability, dNAAT delivers unparalleled precision.
Applications of dNAAT
- The digital molecular analysis approach known as dNAAT uses microscale reactors to “digitize” target molecules in compartments that are only a few microns in size.
- Uniform droplet dispersion and environmental control enhance precision in high-throughput screening, quantitative research, and single-cell or molecule analysis.
- Environmental analysis and single-molecule studies are two areas in which dNAAT finds application.
- More recently, developments in microreactor integration with real-time detection technologies have expanded the field’s prospective uses of dNAAT beyond basic science to include clinical diagnostics.
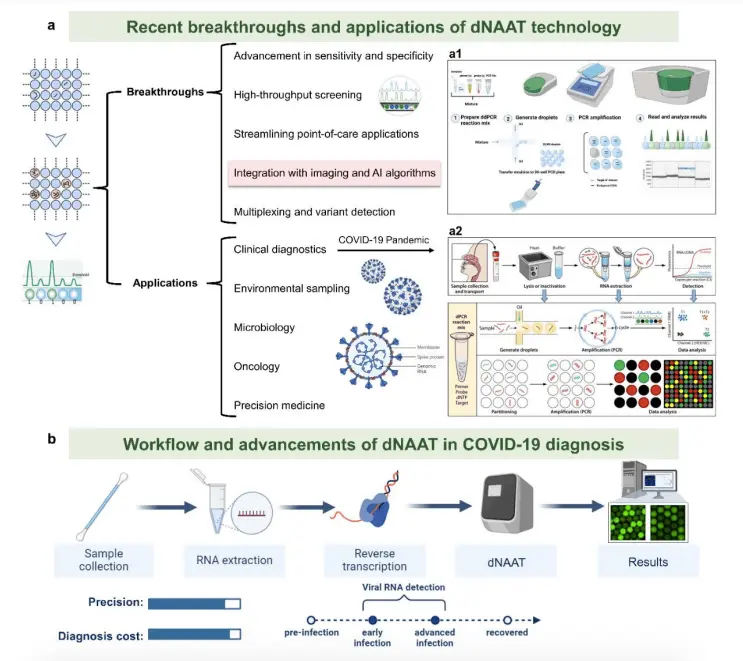
Image Source: https://arxiv.org/abs/2407.21080v1
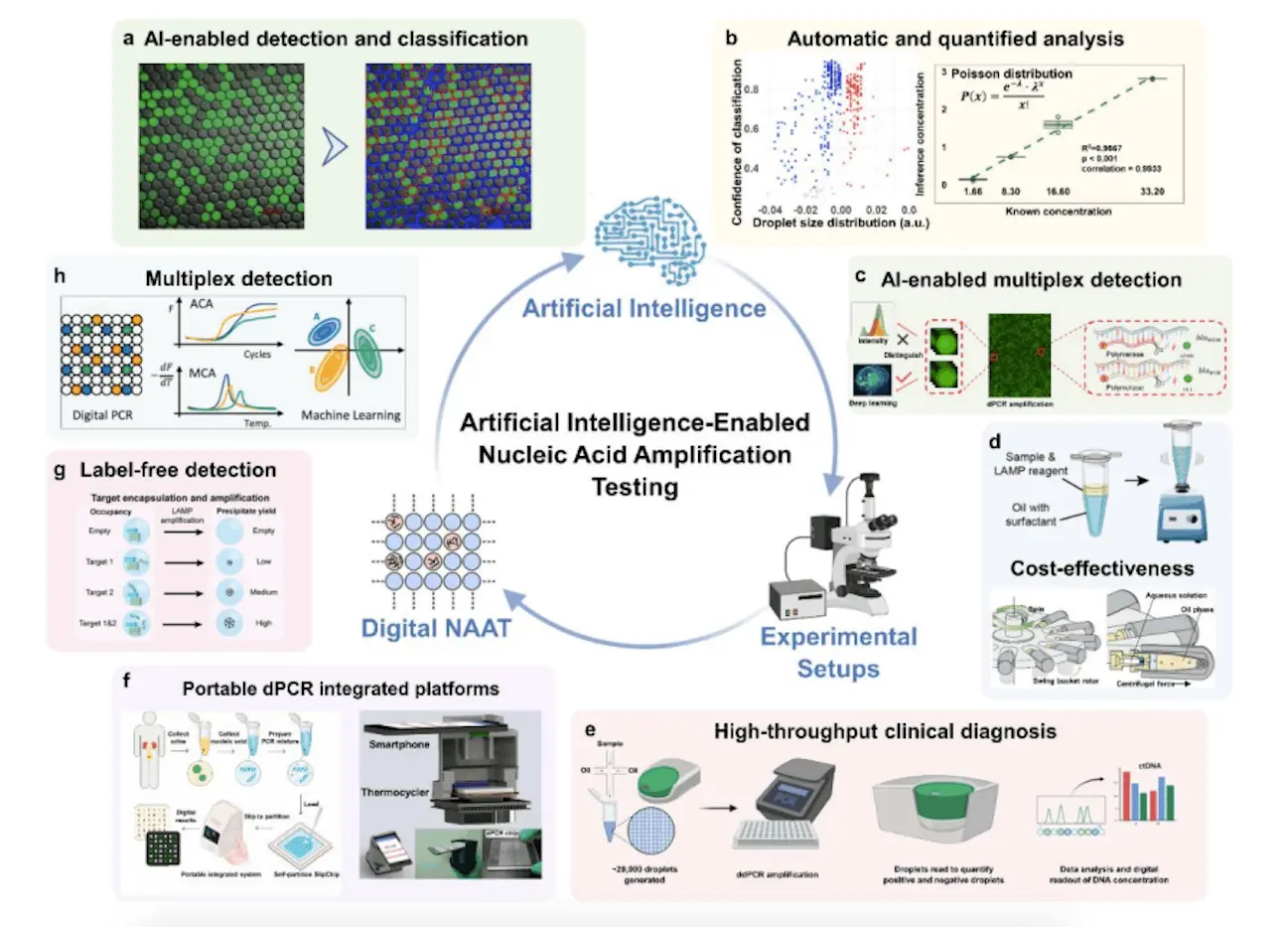
AI and Nucleic Acid Amplification Testing (dNAAT)
The developments in dNAAT (digital nucleic acid amplification testing) have opened the door to possible AI integration. This study looks at Point-of-Care Testing (POCT) technologies and cutting-edge AI-based dPCR and dLAMP detection models. SOTA models achieve fully automated procedures and improved accuracy in decoding dNAAT end-point image information, according to the investigation. Nucleic acid analysis is entering a high-throughput, robust, and affordable era thanks to the incorporation of AI models. The review underscores the significant impact of artificial intelligence on the trajectory of molecular diagnostics.
Conclusion
The study examines the development of dNAAT technologies with an emphasis on how they could revolutionize molecular diagnostics and precision medicine. Invention and early development, commercial development and advancements, integration, and widespread use are the three key stages of research. From Kary Mullis’s first-generation PCR in 1983 to Dr Bert Vogelstein’s conception of dPCR in 1999, the history of PCR technologies is traced to Invention and Early Development. Commercial Development and Advancements is concerned with bringing new technologies to market, such as digital LAMP in 2011 and LAMP in 2000, and incorporating AI to improve capabilities and performance. The expectation is that the general use of these cutting-edge systems will follow the smooth integration of AI optimization with therapeutic applications.
Article Source: Reference Paper
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.