Vision language modeling has been transformed by transformational AI technology called multimodal learning. Technological developments such as multimodal GPT-4V and LLaVA-Med allow for powerful conversational agents that can accomplish tasks without error. However, biomedical settings pose particular difficulties, which is why programs like LLaVA-Med are trying to customize instruction-tuning. The researchers induce significant progress in the field of biomedical technology in this study. Examples of these developments include the creation of the Llama3-Med model, which achieves zero-shot performance on biomedical visual question answering benchmarks, improving performance by over 10% on average, and the introduction of a new instruct dataset enriched with medical image-text pairs from Claude3-Opus and LLaMA3 70B. Medical practitioners now have access to more accurate and dependable tools because of these improvements.
Introduction
Multimodal data integration is achieved using auto-regression in artificial intelligence transformative approaches, e.g., LLaVA and Qwen. These AI initiatives show a noteworthy aptitude for managing intricate inputs, paving the way for creating conversational agents that can accomplish various vision-language activities with a zero-shot approach. By combining textual and visual data in a seamless manner, these models improve AI capabilities in a variety of applications by producing replies that are coherent and contextually relevant.
Even though these developments are encouraging, there are unique difficulties when implementing these technologies in the biomedical industry. More specialized models than those designed for general domains are required due to the particular properties of biomedical image-text combinations. Through the use of large scientific datasets like PMC-15M, recent programs like LLaVA-Med have particularly tailored instruction-tuning for the biomedical environment, improving models’ ability in complex biological visual processing. This technique uses data from PMC-15M image-text pairs to build a wide variety of biomedical multimodal education using GPT-4. A curriculum learning mechanism is then used to fine-tune a vision-language model. These initiatives highlight the potential of tailored multimodal models to satisfy the complex requirements of biomedical applications, indicating a notable advancement in this specialized field.
Key Contributions of this Work
- Firstly, rather than relying only on GPT-4, researchers have created a new training dataset that is enhanced with medical image-text pairs that were jointly produced by the state-of-the-art Claude3-Opus and LLaMA3 70B models. This dataset offers a strong additional resource for model training by greatly expanding the scope of samples beyond those in current collections, like llava-med-instruct-60k. This dataset includes a more representative and diversified mix of textual data and biomedical imaging by improving the diversity and richness of our training materials.
- Second, using MM1, LLaVA-Next, and Mini-Gemini as models, researchers develop a novel picture encoding technique that uses hierarchical representations at different resolutions, ranging from high to low. Given the complex and detailed nature of biomedical photography, this strategy improves the granularity with which medical images are analyzed—a critical improvement. The models can interpret visual input more precisely and deeply by catching little details at different scales. The models can provide more accurate and contextually relevant replies in biological contexts because of this hierarchical encoding method, which guarantees the preservation of important features.
- The Llama3-Med model, which is based on the strong basis given by the well-received open-source large language model (LLM), LLaMA3, is finally introduced by the researchers. It makes use of our enriched dataset and sophisticated encoding techniques. On reputable visual question-answering benchmarks, including VQA-RAD, VQA-PATH, and SLAKE, Llama3-Med has demonstrated state-of-the-art (SoTA) performance never before seen in the biomedical field. This noteworthy accomplishment not only shows how effective our methodological improvements are but also sets a new standard for further study in the same area. With tools that are more accurate, dependable, and skilled at negotiating challenging medical situations, Llama3-Med’s exceptional performance highlights how it has the potential to revolutionize the application of AI in the biomedical industry.
Features of Llama3-Med 8B Model
All datasets and question types show that the Llama3-Med 8B model performs better in zero-shot than other models; the only exception is VQA-RAD, where performance peaked because of insufficient training data. The model’s effectiveness is ascribed to its thorough training on a broad and contextually rich biomedical dataset, which improved its capacity to efficiently generalize to new and unexplored data. For real-world applications, especially in dynamic situations like healthcare, this durability and adaptability are essential. Llama3-Med 8B’s sophisticated model architecture, extensive specialized datasets, and superior image encoding techniques position it as a premier solution for biomedical visual question answering and establish a new standard for future research and development. These findings emphasize how crucial it is to have an adequate and varied training set.
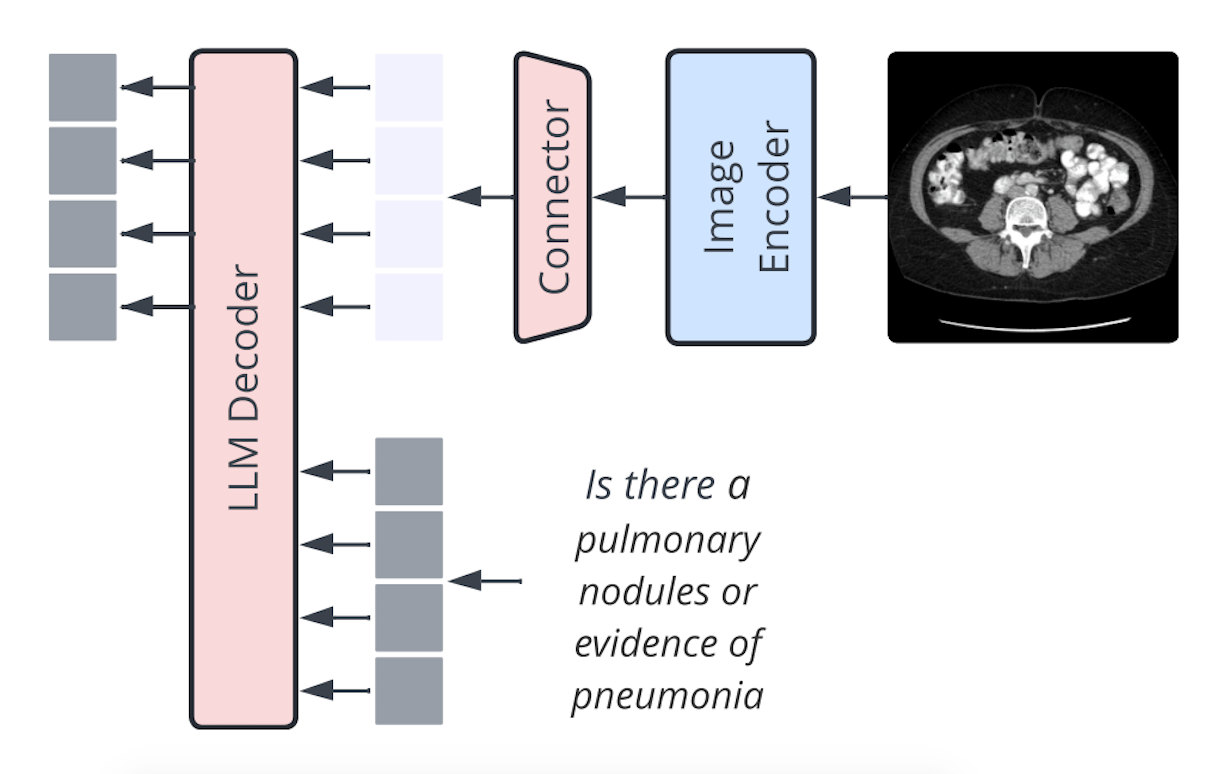
Llama3-Med: Architecture Overview
Llama3-Med is a model architecture that leverages pre-trained language models such as GPT2-XL, BioMedLM, and BioGPT to convert visual characteristics into a visual prefix using a three-layer MLP network. The prefix tuning of language models is comparable to this method, with parameter sizes ranging from 1.5B to 2.7B. The model uses a 7B LM and a linear projection, much like the connector design by Li et al. (2023). Apple researchers found that while the type of VL connector has no bearing, the number of visual tokens and image resolution are the most important factors.
Conclusion
In the realm of biomedicine, the AI-powered Llama3-Med model has shown to be a state-of-the-art tool. It integrates hierarchical representations, vision-language modeling, and a new training dataset that is enhanced with medical image-text pairs. Using this novel technique, the model can interpret intricate biomedical images, giving medical practitioners more precise and contextually appropriate tools. Future research will focus on improving model performance through domain-specific pretraining, ongoing instruction tailoring, and the growth of biological datasets to maximize the benefits of AI in healthcare.
Article Source: Reference Paper | Codes available on GitHub.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.