Scientists from the University of Bayreuth, Germany, introduced ProtGPT2, a language model for protein design trained on the protein space that generates de novo protein sequences following the principles of natural ones. ProtGPT2-sequences predicted using AlphaFold result in well-folded, non-idealized structures. ProtGPT2 generates sequences in a fraction of seconds and has open source access. Novel proteins designed for particular purposes have the potential to solve a wide range of environmental and biological issues.
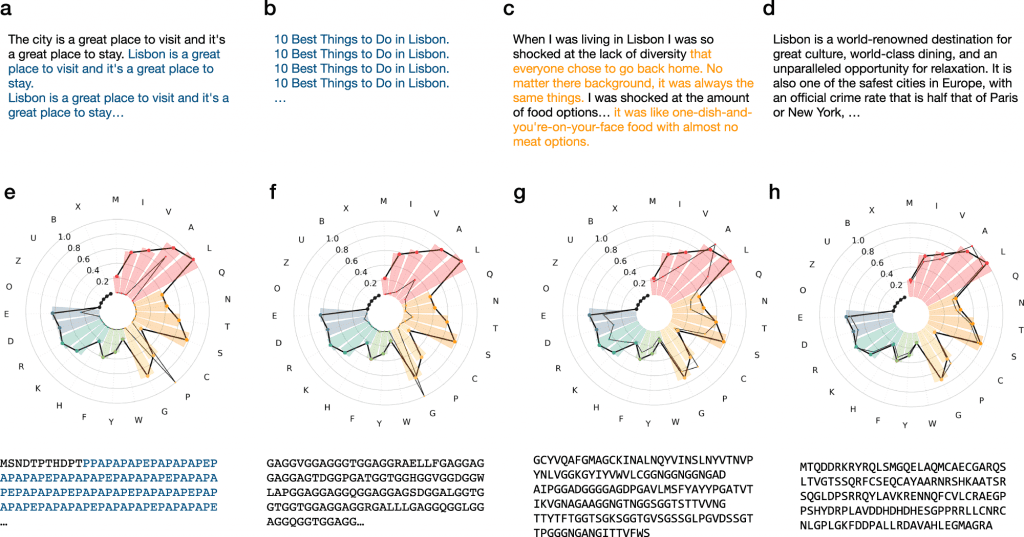
Image source: https://doi.org/10.1038/s41467-022-32007-7
It has long been observed by us that protein sequences are analogous to human languages. As with human languages, protein sequences also consists of letters from a chemically defined alphabet, the natural amino acids, which arrange in order to generate secondary structural elements (or “words”), which then construct domains (or “sentences”) that perform a function ( or “meaning”). During evolution, proteins have undergone point mutations, duplications, and recombinations that have greatly diversified themselves. In spite of significantly divergent sequences, comparisons have been able to detect similarities between the proteins.
The most compelling similarity between proteins and natural languages is that they are information-complete that is, they tend to store the structure and the function in their amino acid order. To analyze protein sequences, several studies have tried to reduce their large dimensionality to a few discernible dimensions. It is possible to represent protein structures using either hierarchical classifications such as the ECOD and CATH databases or Cartesian representations and similarity networks.
In light of the remarkable advances in NLP (Natural Language Processing) finding ways to understand and generate human-like language, we hypothesized that these methods would open up new possibilities for addressing protein-related problems based solely on sequence, such as designing proteins.
A supervised NLP approach involves training input sequences and labels in conjunction to produce a predictive model. In addition to detecting structural similarity, these methods have also been applied to predicting stability. BioSeq-BLM offers an impressive collection of supervised language models for biomolecules. In light of these developments and the growing capabilities of English-speaking models such as the GPT-x series, we set out to develop a generative model capable of:
- Learning protein language.
- Generating fit, stable proteins.
- Understanding how these sequences correspond to natural ones, including whether they are representative of regions of the protein space that we have not observed before.
A versatile tool for language modeling has emerged with the advent of unsupervised learning, where unlabeled data is used for training. TCR-BERT, epiBERTope, ESM, ProtTrans, and ProteinBERT are among the transformer-based models that have demonstrated competitive performance. The majority of these models use BERT-like architectures, and denoising autoencoding training objectives, where the original sentence is corrupted in some way and so is needed to be reconstructed. Another branch of language models can be trained to predict subsequent words given a context, known as autoregressive training. Scaling up unsupervised language models has played a significant role in driving advances in NLP. A language model assigns probabilities to words and sentences based on statistical data.
With 738 million parameters, ProtGPT2, an autoregressive Transformer model, is able to generate de novo protein sequences in a high-throughput manner. An autoregressive language generator can be thought of as the product of conditional next-word distributions and a probability distribution for a sequence. A comprehensive training set of about 50 non-annotated million protein sequences allowed ProtGPT2 to learn the protein language effectively. Protein sequences generated by ProtGPT2 are “evolutionarily” distant from the current protein space while having amino acid and disorder propensities that match natural ones. It is believed that ProtGPT2 is an important step towards efficient high-throughput protein engineering and design because protein design has the potential to solve problems in a variety of fields, including biomedical and environmental sciences.
A majority of the ProtGPT2-generated proteins have natural amino acid properties, while disorder predictions predict that 88% are globular, which is reflective of natural sequences. ProtGPT2 sequences are distantly related to natural ones based on sensitive sequence searches in protein databases, and similarity networks further indicate that ProtGPT2 samples unexplored protein space.

Image source: https://doi.org/10.1038/s41467-022-32007-7
By extending natural superfamilies, ProtGPT2 sequences explore dark areas of the protein space represented by similarity networks. Natural sequences exhibit similar stability and dynamic properties as that of the generated sequences. ProtGPT2 has also successfully retained functional determinants, preserving ligand-binding interaction by superimposing its structure with that of distantly related natural proteins. The ProtGPT2 model was trained on the entire sequence space, so the sequences produced by the model can sample any region, including those known as difficult, such as structures made up of all types of protein molecules and membrane proteins.
Using ProtGPT2 designs, globular proteins can be fitted instantly without further training on a standard workstation. By finetuning the model based on a set of sequences selected by the user, ProtGPT2 can be conditioned to a particular family, function, or fold. The ability of a sequence to fold into an ordered structure is an important feature when designing de novo sequences. It reveals topologies that aren’t captured in current structure databases by AlphaFold prediction of ProtGPT2 sequences, which provides well-folded non-idealized structures with embodiments and large loops.
In ProtGPT2, proteins similar to natural proteins will be screened for new biochemical functions, which can be improved, finetuned, or altered. Extensive screening of ProtGPT2-designed protein libraries might reveal proteins with folds that do not appear in structural databases or functions that do not have a natural counterpart. The ProtGPT2 algorithm efficiently generates sequences that are distantly related to natural sequences but are not acquired through memory and repetition.
A significant step forward towards efficient protein design and generation has been made by ProtGPT2, which prepares the ground for future experimental studies exploring the structural and functional characteristics of designed proteins and their application in the real world. It will be possible to generate specific functions by including conditional tags in future efforts.
Paper Source: Ferruz, N., Schmidt, S. & Höcker, B. ProtGPT2 is a deep unsupervised language model for protein design. Nat Commun 13, 4348 (2022). https://doi.org/10.1038/s41467-022-32007-7
Learn More About Bioinformatics:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Srishti Sharma is a consulting Scientific Content Writing Intern at CBIRT. She's currently pursuing M. Tech in Biotechnology from Jaypee Institute of Information Technology. Aspiring researcher, passionate and curious about exploring new scientific methods and scientific writing.












