CancerGPT, an advanced machine learning model introduced by scientists from the University of Texas and the University of Massachusetts, USA, harnesses the power of large pre-trained language models (LLMs) to predict the outcomes of drug combination therapy on rare human tissues found in cancer patients. The significance of this innovative approach becomes even more apparent in medical research fields where data organization and sample sizes are limited. CancerGPT could pave the way for significant advancements in understanding and treating rare cancers, offering new hope for patients and researchers alike.
Drug combination therapy for cancer is a promising strategy for its treatment. However, predicting the synergy between drug pairs poses considerable challenges due to the vast number of possible combinations and complex biological interactions. This difficulty is particularly pronounced in rare tissues, where data is scarce. However, LLMs are here to address this formidable challenge head-on. Can these powerful language models unlock the secrets of drug pair synergy in rare tissues? Let’s find out!
What are Large Pre-trained Language Models (LLMs) and their Role in Biology?
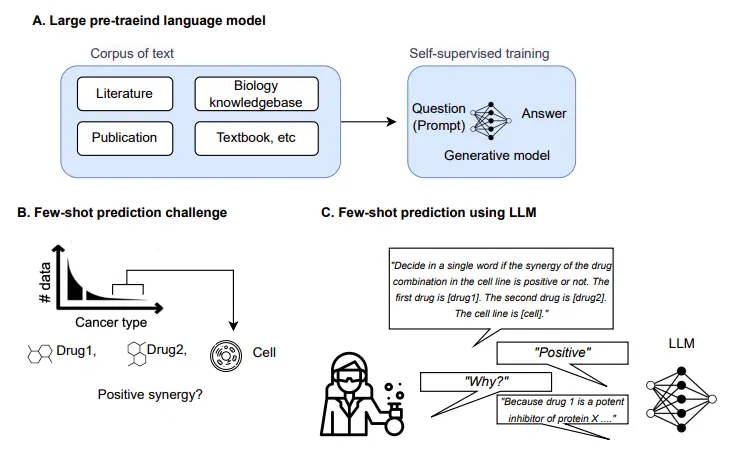
LLMs are a type of artificial intelligence (AI) model that has been trained on massive datasets of text and code. This training allows LLMs to learn the statistical relationships between words and phrases. LLMs have revolutionized the field of artificial intelligence, enabling them to excel in various tasks without task-specific training.
LLMs, such as GPT-3 and GPT-4, has been a game changer in foundation AI model. Few-shot learning, a subset of LLM capabilities, allows these models to tackle new and unseen tasks with minimal or no training data. While LLMs have shown remarkable potential in numerous fields, their applicability in complex domains like biology remains unexplored.
In the context of biology, LLMs hold immense promise as a valuable tool for inferring biological reactions and making predictions, especially in cases where structured data and sample sizes are limited. LLMs can become an innovative approach for addressing biological prediction tasks by leveraging prior knowledge from vast amounts of unstructured literature. The ability to learn from diverse sources of information allows LLMs to capture intricate relationships and patterns within biological datasets, even with minimal training data.
One pressing biological prediction task that could significantly benefit from LLMs is drug pair synergy prediction in understudied cancer types. The experimental data for certain cancer types, like bone and soft tissues, is scarce, making traditional machine-learning models impractical.
LLM-based Approach to Predicting Drug Pair Synergy
To predict whether a drug pair exhibits a synergistic effect in a specific cell line, with a focus on rare tissues that have limited training samples, the researchers propose a novel approach that leverages large pre-trained language models and transforms the prediction task into a natural language inference problem.
The LLM-based prediction models used in the research are GPT-2, GPT-3, and a tailored model called CancerGPT. Using the standard fine-tuning process, these models are fine-tuned using data from common tissues first. Then, they are further fine-tuned with k shots of data from each rare tissue using the few-shot learning approach.
For CancerGPT, the researchers first fine-tuned GPT-2 with a large amount of common tissue data to learn relational information between drug pairs in these tissues, similar to collaborative filtering. This approach assumes that certain drug pairs exhibit synergy regardless of the cellular context, allowing to predict synergy in new cell lines from different tissues. Additionally, the researchers incorporated information on the sensitivity of each drug to the given cell line, using relative inhibition scores as a measure of sensitivity. As a result, the connection between drugs and cell lines was better understood in more detail.
CancerGPT’s Performance and Evaluation
The LLM-based models (CancerGPT, GPT-2, GPT-3) and baseline models (XGBoost, TabTransformer) were evaluated using AUROC and AUPRC metrics, commonly used to assess binary classification performance. LLM-based models achieved comparable or better accuracy in most cases than baseline models. CancerGPT and other LLM-based models (GPT-2, GPT-3) accurately predicted drug pair synergy in rare tissues with limited training data.
The accuracy of the models varied depending on the tissue types, as each tissue possessed unique characteristics and had different data sizes. CancerGPT and GPT-3 exhibited higher accuracy with more shots in specific tissues like the endometrium, soft tissue, and liver, indicating that a few shots of data complemented the prior knowledge encoded in these models. However, additional training data did not always improve the LLM-based models’ performance in certain tissues, like the stomach and urinary tract.
Comparing LLM-based models, CancerGPT and GPT-3 generally outperformed GPT-2, with GPT-3 showing higher accuracy in tissues with limited data or unique characteristics. The experiment on LLM’s reasoning demonstrated that it could provide biologically plausible justifications for its predictions backed by scientific literature. However, some predictions lacked existing scientific evidence, implying the LLM’s capability to infer unseen synergistic effects by combining known scientific facts.
The Promise of CancerGPT
CancerGPT can be used to predict drug pair synergy in rare tissues with minimal data and features. This breakthrough could have significant implications for drug development for understudied cancer types. The approach could significantly advance computational biology, where obtaining abundant training data is not readily possible.
The LLM-based few-shot prediction approach could be applied to a wide range of diseases beyond cancer. For instance, this approach could be used in infectious diseases, where the prompt identification of new treatments and diagnostic tools is crucial. LLMs could help researchers quickly identify potential drug targets and biomarkers for these diseases, resulting in faster and more effective treatment development.
Limitations of the Model
Despite aiming to demonstrate the potential of LLMs as a few-shot prediction model in biology, the present study has certain limitations:
- The LLMs were evaluated only on a few biological prediction tasks. This implies that how generalizable they are to other tasks needs to be elucidated. For example, the study did not assess LLMs on drug discovery or disease diagnosis tasks.
- The study lacks an investigation into how the information gleaned from LLMs complements existing genomic or chemical features. This is an important aspect, as it is likely that LLMs will be most useful when they are coupled with other sources of information.
- The study did not verify the accuracy of GPT-3’s arguments. This is a critical limitation, as it is possible that GPT-3 could make inaccurate or misleading predictions.
- LLMs can also contain biases that humans have. This is a potential problem, as it could lead to LLMs making biased predictions.
Conclusion
CancerGPT represents a pioneering step in utilizing large pre-trained language models for few-shot drug pair synergy prediction in rare tissues. The model’s accuracy and adaptability hold promise for improving drug development in cancer types where data scarcity is a significant challenge. The study highlights the broader applicability of LLMs in biology, enabling quicker identification of potential treatments and diagnostic tools. With further advancements in language model technologies, LLMs have the potential to make even more significant contributions to the field of biology in the future.
Article Source: Reference Paper
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.