Scientists from the Microsoft Research Lab, Cambridge, have designed a Self-Supervised Neural Network Masked Language Model named BiGCARP to accelerate the identification and classification of Biosynthetic Genome Clusters (BGC), which are responsible for generating Natural Products essentials for many industries. This is the first evidence of using Pfam domains as tokens in the Language model. BiGCARP achieves state-of-the-art accuracy in product class prediction.
Introduction to Natural Products
Growth essential substances like Protein, Carbohydrate, Lipid, Nucleic Acids, etc. are called Primary metabolites. On the other hand, Secondary metabolites are not essential for growth but are synthesized by bacteria, fungi, plants, and some marine animals as part of survival strategy and adaptability. These structurally and chemically diverse specialized bioactive molecules are termed Natural Products (NP) and have broad spectrum applications in agriculture (e.g., herbicide, pesticide, insecticide) and the pharmaceutical (e.g., treatment of infectious diseases, tumors, cardiovascular diseases, Immunosuppressive, etc.) industries. Approximately 60% of approved small-molecule medicines are related to natural products. Typically all microbes synthesize small bioactive molecules for nutrient acquisition, communication, and, most importantly, chemical warfare against neighbors. These compounds have already proved their potency as the scaffold of therapeutic leads for drug targets. For instance, who hasn’t taken antibiotics like Erythromycin once in life? It is a semisynthetic Natural product obtained from Actinobacteria Streptomyces.
Nonetheless, the Healthcare system still demands new discoveries and designs of Natural Products as therapeutics. Prior to the Next Generation sequencing and genomics, the search for natural products from microorganisms was conducted primarily using culture-dependent techniques, which overall include the following steps:
- Fermentation.
- Testing of fermentation broths.
- Phenotypic screening to check the effect of the NPs on the different cell lines and compound isolation.
The issues are that only 1% of microorganisms are culturable in the laboratory, and on top of that, in laboratory conditions, most of the secondary metabolic pathways remain silenced. The discovery of biosynthetic genes encoding natural products has directed the way toward unraveling fundamental facets of biosynthesis and the discovery of new drug leads. Sequencing the Streptomyces genome in the 80s was the beginning of this understanding.
Genomic Exploration and Identification of BGCs for Natural Product Discovery
Biosynthetic Genome Clusters (BGC) are physically clustered groups of two or more genes in a particular genome that encode enzymes of a biosynthetic pathway to produce a specialized metabolite (Natural Products). The investigation of BGCs will certainly accelerate the discoveries of novel biosynthetic mechanisms and Natural Products. The astounding structural and chemical complexity and diversity of Natural products are now attributed to the assembly-line enzymology model. These biosynthetic systems retain multimodular architecture. Each module constitutes several domains that collectively catalyze rounds of elongation and chemical modification of Natural Products. For example, Polyketide synthases (PKS) and nonribosomal peptide synthases (NRPS) are two major biosynthetic machinery in microbes. The diversification of Natural products certainly bears evolutionary significance, i.e., microbes need to defend themselves from dynamic ranges of surroundings. However, the biosynthetic capacity of microbes and underlying intricate evolutionary processes are yet to be explored.
The most convenient and widely used computational biology method to recognize these natural products is genome mining. The term “genome mining” is now associated with bioinformatics examination tools to detect biosynthetic pathways of bioactive natural products and their functional and chemical interactions. Genome mining involves the identification and prediction of previously uncharacterized natural product biosynthetic gene clusters. The current most extensive collection of mined gene clusters is the “Atlas of Biosynthetic Gene Clusters,” a component of the “Integrated Microbial Genomes” Platform of the Joint Genome Institute (JGI IMG-ABC).
The Genomic studies inferred an interesting fact that microbial genomes possess way more amount of BGCs than was characterized before. NGS, metagenomics, and single-cell sequencing technologies have presented adequate genomic data for innumerable microorganisms. Therefore, the window for identifying secondary metabolite pathways has elevated. The designation of BGCs directly from genomic sequences is paramount in navigating and nominating new natural products.
BGC Recognition Resources
The following resources are utilized commonly to designate Biosynthetic Gene Clusters in Genomes:
- antiSMASH (ANTIbiotics & Secondary Metabolite Analysis SHell): a web server enabling gene cluster identification and compound analysis. It uses a set of curated profile-Hidden Markov Models (pHMMs) to call biosynthetic gene families and a set of heuristics to tag a portion of a genome as a BGC.
- ClusterFinder: The algorithm uses a Hidden Markov Model that switches between BGC and non-BGC analysis to look for patterns of broad gene functions rather than searching for specific individual signature genes.
- “Prediction Informatics for Secondary Metabolomes” (PRISM): an open-web tool that connects genomic data, chemical structures, and resistance genes and predicts secondary metabolomes.
- DeepBGC: It utilizes a deep learning genome-mining strategy for biosynthetic gene cluster annotation that addresses the limitations of antiSMASH and ClusterFinder. It uses sets of curated pHMMs to call biosynthetic gene families, and a supervised neural network predicts BGC boundaries and annotates BGC function.
BiGCARP: The Recent Advancement
HMM-based algorithms are unable to recognize higher-order dependencies between genes. Rule-based methods fail to generalize well to new BGC classes. Although DeepBGC demonstrates optimistic improvements in the identification of BGCs in microbial genomes, it is trained on a small number of high-quality annotations. The quality of the predictions is likely to depend on the quality of the negative examples.
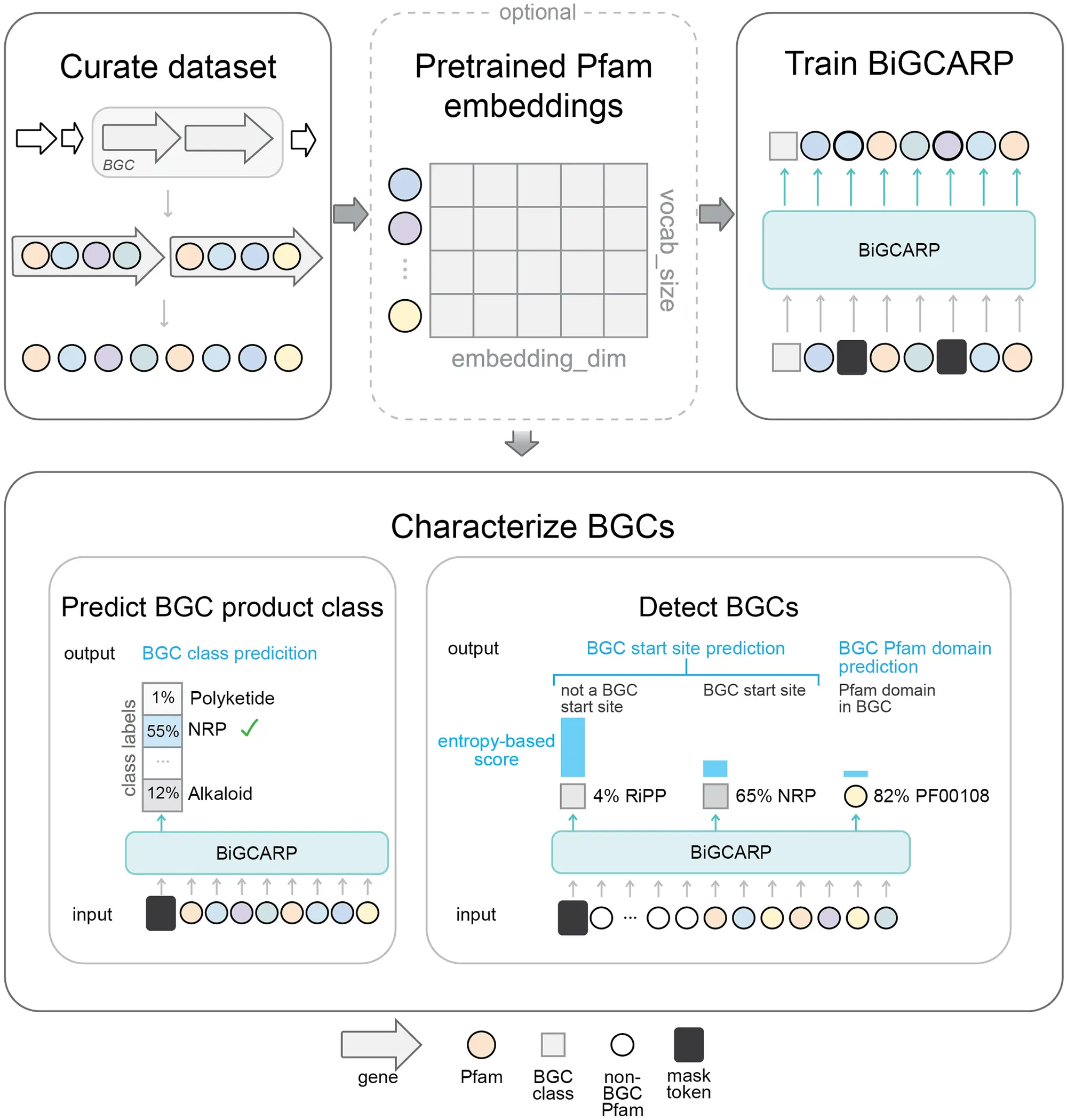
Recently, self-supervised masked language models of biological sequences have been used to inspect biomolecules like protein, DNA, RNA, and glycans. The researchers of Microsoft Research lab applied Self-supervised masked language models to recognize biologically-relevant patterns directly from a large set of BGC examples.
The architecture for the model is based on Convolutional Autoencoding Representations of Proteins (CARP) which is a masked language model of proteins. Therefore, the model is named Biosynthetic Gene CARP (BiGCARP). It learns representations of BGCs based on their Pfam domains, detects & characterizes BGCs, and predicts their product classes.
Conclusion
The BiGCARP model could be applied for downstream analysis like chemical product structure characterization and BGC mining. Nonetheless, the development of a novel natural product certainly requires the collaborative efforts of researchers and industries. The quick adaptation of researchers to the advancement of such technologies will determine the acceleration of new findings.
Article Source: Reference Paper
Learn More:
Aditi is a consulting scientific writing intern at CBIRT, specializing in explaining interdisciplinary and intricate topics. As a student pursuing an Integrated PG in Biotechnology, she is driven by a deep passion for experiencing multidisciplinary research fields. Aditi is particularly fond of the dynamism, potential, and integrative facets of her major. Through her articles, she aspires to decipher and articulate current studies and innovations in the Bioinformatics domain, aiming to captivate the minds and hearts of readers with her insightful perspectives.