In the world of molecular design that is constantly evolving, it is always essential to ensure that along with the structure and properties of newly designed compounds appeal to synthetic chemists; there are ways to synthesize the compound. The ability to have cost-aware decision-making algorithms is a novel opportunity with a focus on the qualified creation of molecules limited by the given costs. A new algorithm known as SPARROW (Synthesis Planning and Rewards-based Route Optimization Workflow) was developed by researchers at MIT, Cambridge. Intended as a breakthrough on the way to achieving the goals of greater efficiency of the molecular design cycles and reduction of the overall synthetic cost, the tool in question seems to have done its job right.
Computational Workflow Limitations
Although computational workflows aid in ranking molecules based on the number, they involve several assumptions on testing costs and potential applications. Presumably, some aspects of generative models may offer molecules that are scientifically possible to synthesize. Therefore, it has high evaluation costs. Substitution based on synthetic complexity or Synthetic Accessibility Score independently improves but does not consider the sum of costs associated with batch synthesis that may hinder a holistic approach to molecular selection.
The SPARROW Solution
Because of optimizing the nanoscale design, the SPARROW algorithm comes to the rescue when tackling problems of higher order. It remarkably balances the growth of predicted properties with forward synthesis and backward analysis, maintaining the cost of synthesized chemicals in line with its expected information value. In this way, utilizing case studies, SPARROW demonstrates learning non-additive costs, integrating common intermediates, and merging the approaches from library- and de novo-based design to fundamentally transform the molecular design optimization space.
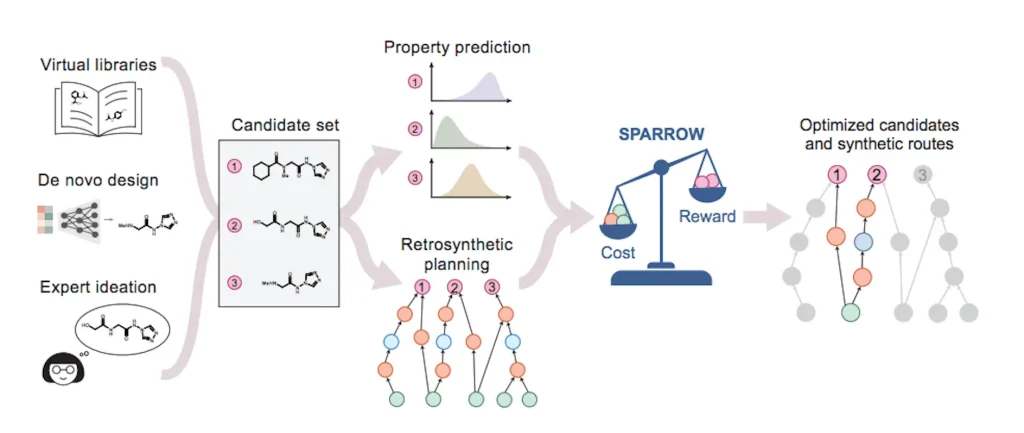
Methodology Used
The type of activity used is in the formulation of the specific SPARROW algorithm by considering the cost factors. This algorithm uses molecular reconstruction, molecular design, and property prediction to select molecules. They use examples to illustrate how the described algorithm can help optimize the best/worst-case costs versus their utility in molecular design.
- Candidate design space: Candidate design space can be defined as the retrosynthetic graph, namely, the graph that contains candidate molecules and the reactions that create them. This graph can be drawn in the form of a directed bipartite graph differentiating between the nodes representing reactions and the nodes representing the compounds, in which the relational schema is the reactants and products. It provides a map through which one is likely to examine and comb the possible pathways of synthesizing a molecule or compound.
- Decision variables and constraints: The decision variables of the SPARROW optimization process are binary variables that relate to the compound as well as reaction nodes, with the two sets being as follows: these variables incur some constraints that ensure that the route selection starts with appropriate material while also ensuring the selection of parent nodes in addition to avoiding synthetic cycles sourced from compounds that cannot be bought. The constraints are also essential to the modeling process as they ensure that the predicted synthetic strategies are efficient and financially practicable.
- Linear objective function: The linear objective function in SPARROW is to select the strategy path to accomplish more cumulative rewards while avoiding more synthetic costs and risking failure of the reaction to the strategy. This optimally serves utility and cost requirements by ranking the candidate reactions according to high returns, high-score reactions, and catalysts composed of few and cheap raw materials. This function is scalarized to facilitate the optimality assessment of the solutions through coding the objectives so that solutions with improved objective function, utility value, or affordability can be selected.
- Optimization Solver: OF1 follows the linear optimization theory, and SPARROW formulates a linear optimization problem. The optimization model is solved using PuLP and the open-source Cutting plane branch and check (CBC) solver. The solver works with certain tolerances or thresholds in order to have convergence and compute precise values for the realized pathways.
- Baseline: In the analysis of the results, some baselines were also used for comparative mechanisms of the work. Three selection strategies relied on reward, synthetic accessibility score, and other scores based on both of these parameters. From here, the routes were ranked by their plausibility score, especially by looking for starting inputs that are accessible and could be bought.
Optimizing Large Candidate Sets
A fact that stands out in SPARROW is how effectively the function can compress time to find an optimal route for numerous candidates. Thus, in addition to the optimization of 300 alectinib analogs, SPARROW was also able to identify novel synthesis routes to 215 targets with significant overlapping reaction steps, proving that it had established a higher level of efficiency in route optimization.
Cost Analysis Validation
To strengthen its argument and to prove that it is on the right track in achieving its vision, SPARROW conducted a detailed cost analysis. Comparing compound prices from October 2023 to March 2024 would successfully prove the efficiency of SPARROW in sorting baseline measures, indicating its accuracy in controlling synthetic costs.
Conclusion
The incorporation of cost-conscious decision-making into molecular design marks a significant development in the area. This algorithmic framework, which combines computational power, machine learning, and economic concerns, provides a complex tool for researchers and industry personnel. It has the potential to not only accelerate invention but also ensure that novel molecular designs are scientifically revolutionary and commercially feasible. As this technology advances, it will play an increasingly important role in influencing the future of molecular design and its applications across a wide range of sectors.
Article Source: Reference Paper | Reference Article | SPARROW is open source and is available on GitHub.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.
















