Scientists from Aalto University and the University of Luxembourg have developed a machine learning model that can accurately identify small molecules. The model has the potential to be used in various fields, including medicine, drug discovery, and environmental chemistry. The model was trained using data from numerous laboratories and is now considered one of the most reliable tools for identifying small molecules.
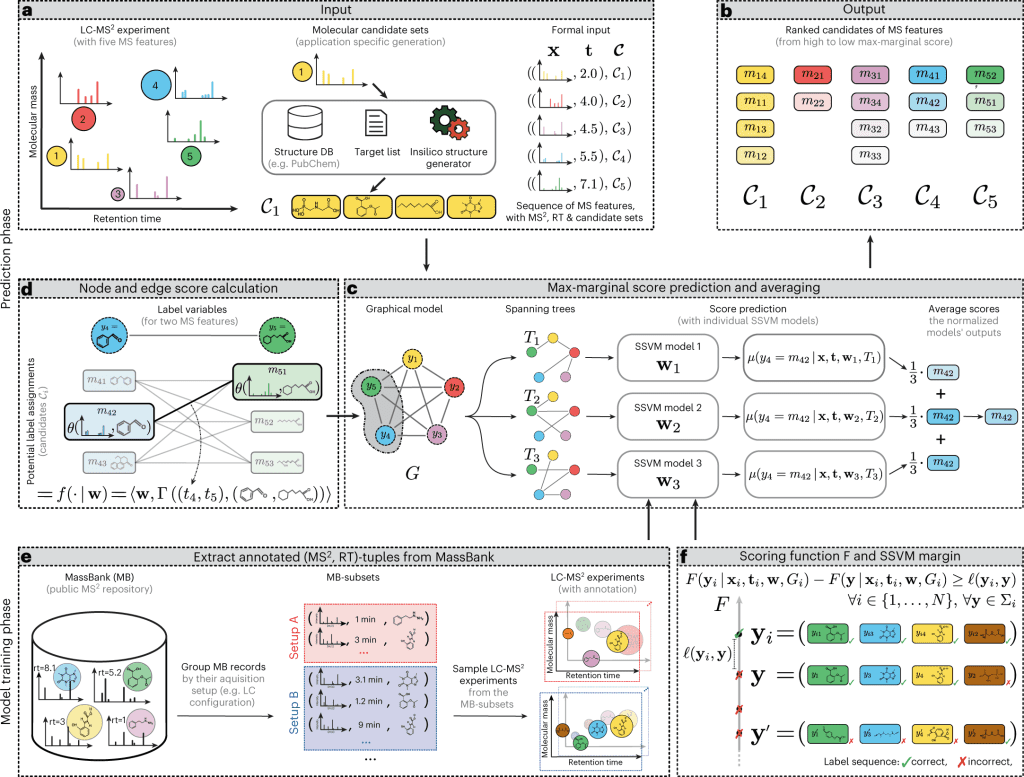
Image Source: https://doi.org/10.1038/s42256-022-00577-2
There are thousands of different small molecules called metabolites in the human body, which play crucial roles in transporting energy and transmitting cellular information. These molecules, which are found in all cells and tissues, are difficult to distinguish from one another in a blood sample analysis due to their small size. However, identifying these molecules is important for understanding how factors such as exercise, nutrition, alcohol consumption, and metabolic disorders can impact overall health and well-being.
Metabolites are typically identified using a separation technique called liquid chromatography followed by mass spectrometry. This technique involves running a sample through a column, which separates the metabolites based on their flow rates (or retention times). Mass spectrometry is then used to further refine the identification process by sorting the metabolites based on their mass. Additionally, researchers can use a technique called tandem mass spectrometry to break down the metabolites into smaller pieces and analyze their composition. These techniques are commonly used to identify and characterize metabolites in order to understand their roles in the body and how they may be affected by various factors.
According to Professor Juho Rousu of Aalto University, even with the most advanced techniques, it is still difficult to identify more than 40% of the molecules present in a sample without making assumptions about the characteristics of the candidate molecules. This means that there is still a significant amount of unknown or unidentified molecules that remain to be discovered and characterized. This can be a challenge for researchers who are studying the roles of small molecules in various biological processes and seeking to understand how they are affected by different factors.
By using machine learning techniques, scientists can gain a deeper view of these small molecules and their roles in the body, which can help improve our understanding of how different factors affect human health. This information can help in the development of treatments and therapies for various conditions that are related to metabolism and cellular function.
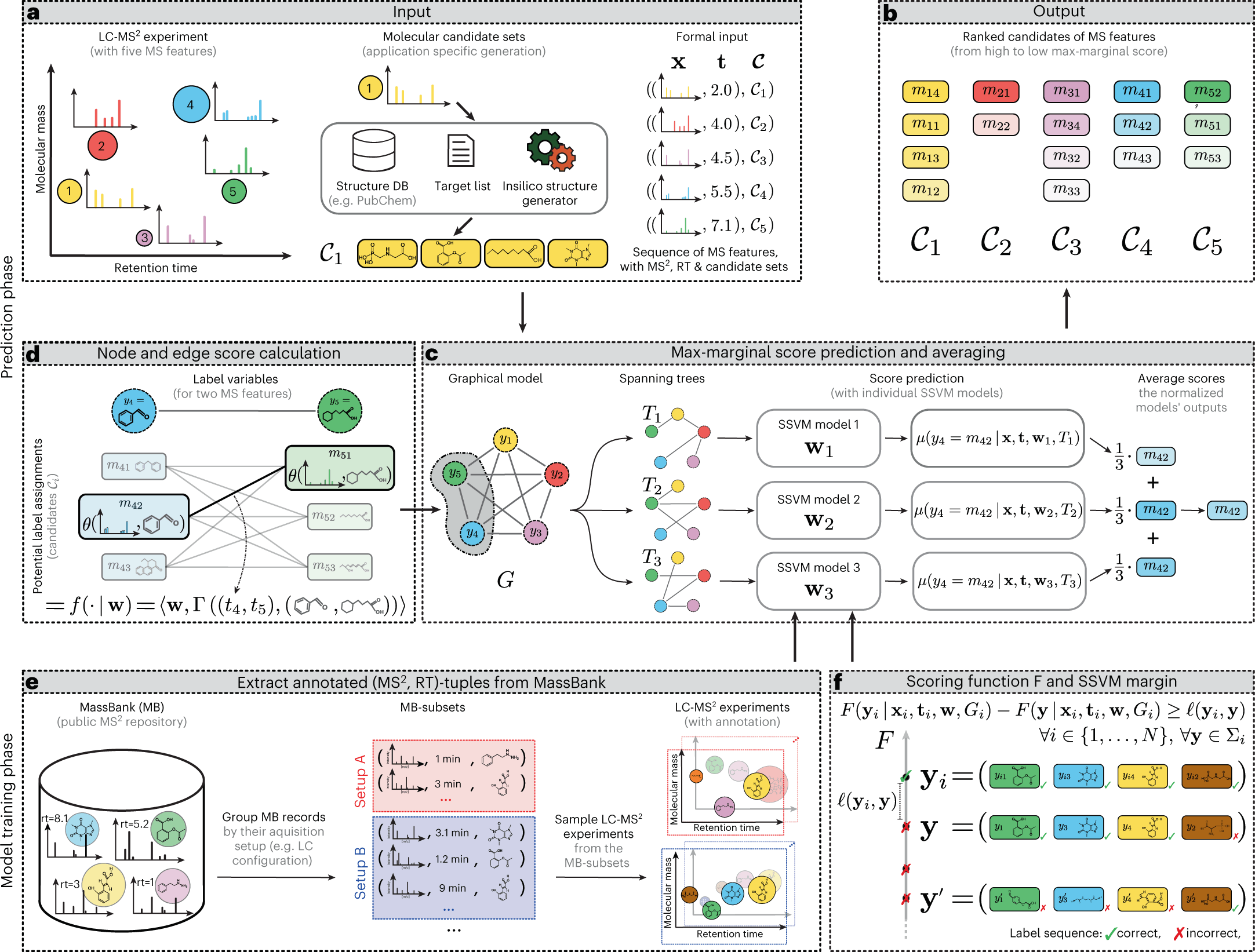
A new machine learning model, “LC-MS2Struct,” for identifying small molecules has been developed by a group led by Prof. Rousu. This model was recently published in the journal Nature Machine Intelligence. The model is innovative and potentially useful for studying small molecules and their roles in various biological processes.
The newly developed machine learning model for identifying small molecules offers researchers a more comprehensive view of these molecules and their roles in various biological processes. It is an open-source model with the potential to facilitate research into the identification and treatment of metabolic disorders, such as diabetes, as well as cancer and other diseases. The model may help researchers develop new therapies and treatments for these conditions by providing a more detailed understanding of small molecules.
The machine learning model for identifying small molecules has been trained using data from numerous laboratories worldwide, contributing to its accuracy. One of the standout capabilities of this model is its ability to distinguish between mirror-image molecules, known as stereochemical variants. Previous identification tools have not been able to differentiate between these variants, but the new model’s ability to do so is expected to open up new possibilities in drug design and other fields. This is a significant advancement in the field of chemistry and has the potential to lead to important discoveries and developments.
Final Thoughts
In conclusion, the use of machine learning in the study of small molecules is providing scientists with new insights and capabilities and is helping to advance the field of chemistry. Small molecules, also known as metabolites, play critical roles in various biological processes, including the regulation of gene expression, cell signaling, and metabolism. However, these molecules are often present in complex mixtures, making it challenging to identify and quantify them. Using techniques such as mass spectrometry and predictive modeling, scientists can analyze large amounts of data and gain a deeper understanding of small molecules. This information can be used to improve our understanding of how different factors, such as exercise, nutrition, and alcohol use, affect human health and can also be used to design new small molecules for use in drug development.
Article Source: Reference Paper | Reference Article
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.