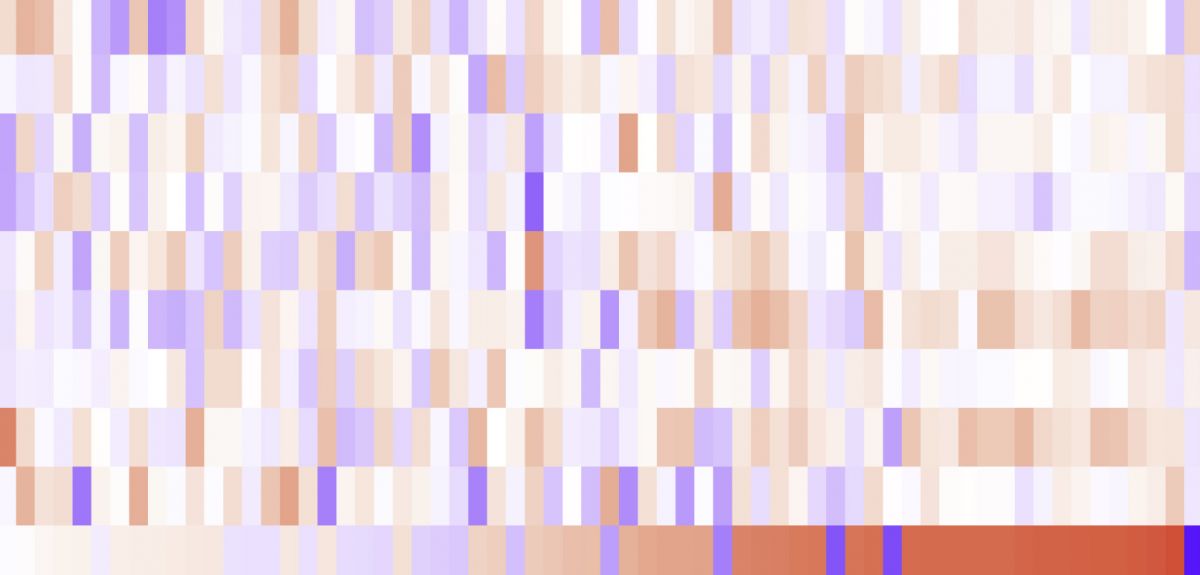
In an integrated comparison with influenza and sepsis patients against healthy volunteers, the researchers show a complete multi-omic blood atlas for patients with different COVID-19 severity. Immune signatures and host response correlations are identified.
Research describing markers of COVID-19 disease severity has been published by a multi-disciplinary, cross-divisional collaborative team led by the University of Oxford. Professor Julian Knight of the Wellcome Centre for Human Genetics led a team of over 200 researchers, who performed multiple ‘omics analyses on the blood of patients with varying COVID-19 severity and compared them to patients with severe influenza, sepsis patients, and volunteers that were healthy.
Image Source: A blood atlas of COVID-19 defines hallmarks of disease severity and specificity.
Cells, their inflammatory mediators, and networks were identified as prospective indicators, including progenitor cells and particular myeloid and lymphocyte subsets, immunological repertoire traits, acute phase response, metabolism, and coagulation. COVID-19 was characterized by persistent immunological activation involving p38MAPK/AP-1. The plasma proteome allowed for sub-phenotyping of patients into clusters that might be used to predict severity and prognosis. The whole dataset’s tensor and matrix decomposition showed feature groups associated with illness severity and specificity. The systems-based integrative approach and blood atlas will inform future medication development, clinical trial design, and personalized medicine methods for COVID-19.
Professor Julian Knight said: ‘This has been possible through an extraordinary team from different disciplines working collaboratively to address an unprecedented health challenge. The work has advanced our understanding of why a small minority of those infected with SARS-CoV-2 develop severe illness, and provides opportunities to identify therapeutic targets and personalise care for patients. An in-depth multi-omic approach revealed the nature of underlying immune dysfunction, the extent to which this is specific to COVID-19 and how this varies between people.’
The multi-omic integrated blood atlas delineates the host immune response in COVID-19 from the beginning of the UK pandemic to clinical trial-led adoption of authorized therapies or immunization. This offers the community a one-of-a-kind reference resource for replication and meta-analysis, as well as an interpretive browser for datasets generated from interventional trials. Patients must be better differentiated according to illness severity, underlying pathophysiology, and infectious etiology using integrative techniques like the one used in the research. This will be critical as we look for new therapeutic targets and the possibility of a precision medicine treatment strategy that is suitably timed and tailored to the people most likely to benefit from a specific intervention.
Story Source: Ahern, D. J., Ai, Z., Ainsworth, M., Allan, C., Allcock, A., Ansari, A., … & Sansom, S. N. (2022). A blood atlas of COVID-19 defines hallmarks of disease severity and specificity. Cell, 185 (5), 916-938. DOI: https://doi.org/10.1016/j.cell.2022.01.012.
https://www.ox.ac.uk/news/2022-03-03-covid-19-multi-omic-blood-atlas-combat-published
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar