Scientists from the National Cancer Institute, Bethesda, and the University of Maryland have developed a computational tool to determine gene-target combinations for personalized cancer therapies. The tool has the potential to aid in the creation of novel combinatorial treatments for cancer patients.
Image Source: The landscape of receptor-mediated precision cancer combination therapy via a single-cell perspective
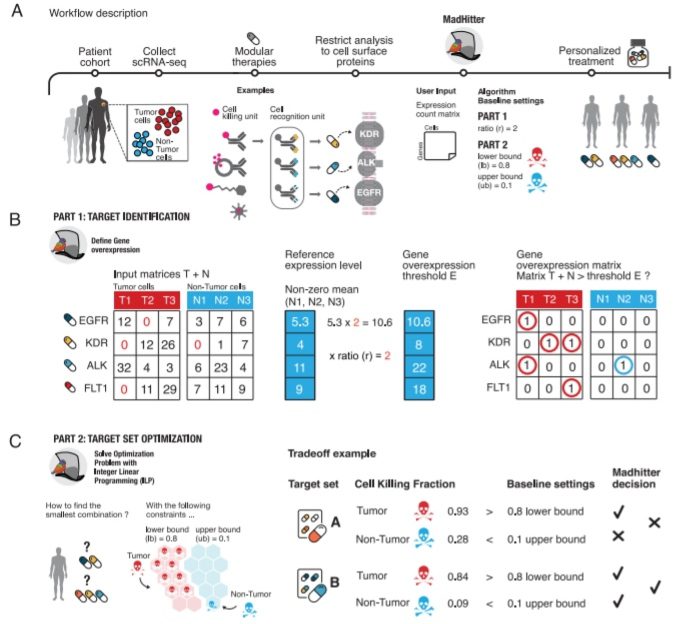
For mining a vast cohort of single-cell transcriptomics information, the scientists utilize combinatorial optimization techniques to chart the landscape of ideal combination therapies in cancer. They assumed that every individual therapy could target any of 1269 genes encoding cell surface receptors, which might be the targets of CAR-T, conjugated antibodies, or coated nano-particle treatments.
The scientists observed that in most cancer types, personalized combinations formed of at most four targets are then adequate for killing somewhere around 80% of cancer cells while saving something like 90% of non-tumor cells in the tumor microenvironment.
Nonetheless, as more stringent and selective killing is required, the number of targets needed ascents quickly. Individual targets include PTPRZ1 for brain, head, neck cancers, and EGFR in multiple tumor types. This study gives a computational estimate of the identity and number of targets required in combination to target cancers selectively and precisely.
Personalized Oncology
Personalized oncology offers trust that every patient’s cancer can be dealt with, given its genomic characteristics. A few preliminary trials have recommended that it is feasible to gather genomic data adequately to illuminate treatment decisions.
Meta-analysis of phase I clinical trials finished during 2011-2013 showed that, in general, trials that utilized molecular bio-marker data to impact treatment plans gave better results than trials that did not. In any case, most precision oncology treatments use only a couple of drugs, and resistant clones often arise, underlining the need to convey personalized medicine as multiple agents combined.
Significant potential chances to join systems biology and the design of nanomaterials have been perceived to deliver medicines in combination to conquer drug resistance and join biological effects.
Future of Precision Oncology Treatments
The design of future precision oncology medicines is a very challenging scenario, given the developing acknowledgment that tumors commonly have extensive intra-tumor heterogeneity (ITH) and hence should be targeted with a combination of medicines to such an extent that a considerable number of tumor cells are hit by something like one medication.
The scientists’ examination depends on two emerging technologies:
- The advancement of single-cell transcriptomics and proteomics estimations from patients’ tumors, which are expected to enter progressively into clinical use, and
- The introduction of “modular” treatments that target explicit overexpressed genes/proteins to perceive cells in a specific manner and afterward use either the T-cell immune response or a lethal toxin to kill the tumor cells preferentially.
They centered their analysis around genes encoding receptors on the cell surface, as these might be precisely targeted by any of no less than six technologies, including CAR-T therapy, immunotoxins ligated to antibodies, immunotoxins ligated to mimicking peptides, furthermore, conventional chemotherapy ligated to nanoparticles. These treatments are completely named “modular,” as they include one section that specifically targets the tumor cell through a gene/protein overexpressed on its surface and another part, the cytotoxic mechanism that kills the cells.
Addressing the Selectivity Problem
To address the selectivity issue, different groups have engineered composite types of CAR-T treatments that execute Boolean AND, OR, and NOT gates that have been tested for combinations of up to three target proteins. In silico strategies centering on AND gate and pairs or trios of targets were introduced, and 2538 likely cell surface proteins that are not receptors were analyzed.
The researchers decided to zero in on the simpler logical OR construction since that can be accomplished not just via CAR-T technology yet can likewise be implemented through other modular receptor-mediated treatment technologies by consolidating multiple single-target treatments, assuming that the composite treatment kills a cell, if any of the single treatments kill the cell.
Conceptually, such a logical OR combination treatment can, in any case, accomplish selectivity by choosing targets, every one of which is communicated on a lot higher proportion of cancer cells than non-cancer cells. One of their key contributions was to show that by utilizing techniques from combinatorial optimization, one could see such successful combinations, including many targets, while past studies were restricted to at most three targets.
Beyond CAR-T, the scientists’ analysis applied to a few additional types of modular treatment technologies that depended rather on receptor-mediated endocytosis (RME) delivering a toxin using a targeted receptor to enter the cell. These RME-based technologies incorporate, for instance, conjugated antibodies and toxin delivering nano-particles.
Like CAR-T, those technologies are known not to downregulate the target receptor. For RME technologies and different technologies that work intracellularly, the scientists anticipated joining modular treatments from one technology with the end goal that all treatments utilize the same toxin or mechanism of cell killing, accordingly alleviating the need to test for interaction impacts between pairs of different treatments.
Strategy Discussion from the Multi-Disciplinary Review
In this multi-disciplinary research, the scientists’ employed strategies from combinatorial optimization to dissect openly accessible single-cell tumor transcriptomics data to chart the landscape of personalized combinations that depend on modular therapies, including CAR-T treatment. They showed that, for most tumors they studied, four modular medications targeting different overexpressed receptors might get the job done to selectively kill most tumor cells while saving a large portion of the non-cancerous cells.
For the more restricted sets of low-expression genes or the 58 receptors with validated ligand-mimicking peptides, a few patients don’t have plausible solutions, particularly as they decreased the TPM expression utilized for filtering the gene set to prevent targeting non-cancerous tissues. These discoveries demonstrate that, on the one hand, scientists designing ligand-mimicking peptides have been keen on choosing targets pertinent to cancer.
Then again, the outcomes propose a requirement for expanding the set of cell surface receptors that can be targeted to enter tumor cells with ligated chemotherapy agents. There are two laid out techniques to distinguish ligand-mimicking peptides called “one bead, one compound” and “phage display.” The two strategies apply to all cell surface proteins, and accordingly, the set of proteins with validated peptides could be extraordinarily extended as nanoparticle-based treatments draw nearer to being utilized in the clinic.
On the preventative side, experiments with target gene sets that were additionally filtered by low expression in normal tissues showed that the individual target set issue could become infeasible in many occurrences. In any event, when the occurrence stayed feasible, optimal cohort treatment set sizes expanded quickly as the expression levels permitted diminished, highlighting potential innate limitations of applying such combination approaches to deal with patients in the clinic and the need to carefully screen their putative wellbeing and toxicity in future applications.
At last, functional enrichment analysis of genes commonly happening in the optimal target sets built up the focal job of the generally studied oncogene EGFR and other transmembrane kinases. Moreover, the researchers observed that the less-studied phosphatase PTPRZ1 is a valuable target, particularly in brain cancer.
The Endpoint
This study utilizes combinatorial optimization tools to examine emerging single-cell data to depict the landscape of possible personalized combinations in cancer medication. The research discoveries uncover promising membranal targets for developing future oncology drugs that might advance the therapy of cancer patient cohorts in a few cancer types. The MadHitter approach and the accompanying software unveiled can be promptly applied to address additional essentially related research questions and analyze extra cancer data sets as they become available.
Story Source: Ahmadi, S., Sukprasert, P., Vegesna, R. et al. The landscape of receptor-mediated precision cancer combination therapy via a single-cell perspective. Nat Commun 13, 1613 (2022). https://doi.org/10.1038/s41467-022-29154-2
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Tanveen Kaur is a consulting intern at CBIRT, currently, she's pursuing post-graduation in Biotechnology from Shoolini University, Himachal Pradesh. Her interests primarily lay in researching the new advancements in the world of biotechnology and bioinformatics, having a dream of being one of the best researchers.