PGDx Elio tissue complete, which the FDA has approved for the analysis of 505 cancer-related genes, will enable the development of more effective precision oncology treatment options.
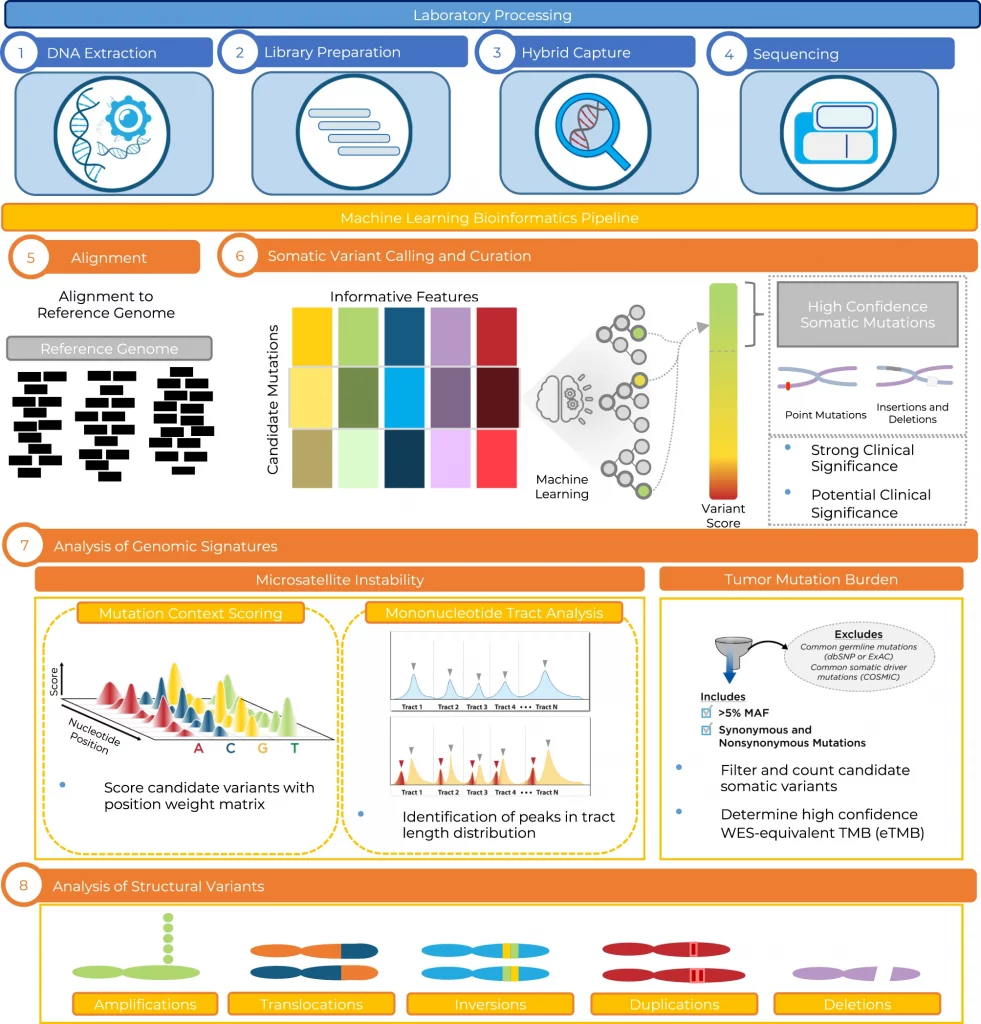
Image Source: Automated next-generation profiling of genomic alterations in human cancers
The researchers from Personal Genome Diagnostics Inc. (PGDx) explain the developmental and analytical validation processes of the PGDx elio tissue complete test. It is an automated NGS-based approach for the comprehensive profiling of genomic alterations in human cancers.
The shortfall of invalidated, distributed, extensive genomic profiling assays for patients with cancer inhibits access to precision oncology treatment.
To address this, the scientists portray elio tissue complete, which had been FDA-cleared for assessment of 505 cancer-associated genes. Independent examinations of clinically and biologically pertinent sequence changes across 170 clinical tumor samples by the utilization of MSK-IMPACT, FoundationOne, and PCR-based strategies uncovers a positive percent agreement of >97%.
The researchers noticed a high concordance with whole-exome sequencing for the assessment of tumor mutational burden for 307 solid tumors (Pearson r = 0.95), and a comparison of the elio tissue complete microsatellite instability detection approach with an independent PCR measure for 223 samples show a positive percent agreement of close to 100%.
At last, the assessment of amplification and translocations against DNA-and RNA-based approaches displays >98% negative percent agreement and positive percent agreement of 86% and 82%, respectively. These strategies give a way to deal with pan-solid tumors with complete genomic profiling with high analytical performance.
The Critical Identification of Genetic Landscape of Each Patient’s Cancer
High-intricacy and extensive next-generation sequence analyses are changing the diagnostic scene of oncology.
Various targeted therapies against proteins impacted by genetic alterations have been more secure and successful than traditional chemotherapies when utilized in a suitable patient population.
This has been effectively shown for various therapeutics targeting the protein products of explicit genes that are changed in human cancer, including the utilization of therapeutics like the following:
- Imatinib in chronic myeloid leukemias carrying BCR-ABL fusion,
- Trastuzumab in ERBB2 (HER2/neu) amplified breast cancer and
- Vemurafenib in BRAF-mutated melanoma
Molecular modifications have likewise been displayed to make a predictive or prognostic impact, like the poor response to anti-EGFR (epidermal growth factor receptor) monoclonal antibodies in patients with mutations at codons 12 and 13 of KRAS in colorectal cancer.
At last, the recently established association between microsatellite instability (MSI) or high tumor mutation burden (TMB) across various solid tumor indications and solid patient response to immune checkpoint inhibitor therapies requires extra testing for these genomic signature biomarkers.
Since the mutations and mutational cycles driving every tumor might be unique, identifying the genetic landscape of every patient’s cancer is fundamental for developing a treatment plan that exploits the developing number of targeted and immune therapies.
Overcoming the Barriers to Broad Diagnostic Analyses
By and large, the way to deal with testing for the presence of targetable mutations or genomic signatures has required multiple single-analyte immunohistochemistry or polymerase chain reaction examinations that frequently test for one alteration at a time.
As guidelines suggest testing for more than ten alterations for non-small cell lung cancer patients, the accessible cancer tissues are in many cases exhausted before significant actionable targets are identified.
Without completely testing possibly significant actionable alterations, a few patients that might have been candidates for explicit therapies would be left without these treatment choices.
This boundary to comprehensive diagnostic analyses can be overcome with the utilization of comprehensive genomic profiling, where different noteworthy actionable sequence mutations, structural variations, and genomic signatures are assessed immediately from a singular tissue specimen.
By eliminating the requirement for multiple tests, more patients might be evaluated for known alterations connected to a targeted or immunotherapy, consequently extending patient access to steadily developing quantities of precision medicine therapies.
An NGS-Based Targeted Assay
While comprehensive genomic profiling has been accessible for quite a while in several specialized laboratories, it has not yet accomplished far-reaching clinical adoption because of an absence of validated assays that could be scaled in local labs and lacking reimbursement connected with regulatory clearance.
Even though a few labs might be fit for developing an NGS-based targeted assay, comprehensive genomic profiling investigations are challenging to develop due to the sophisticated bioinformatics examination and interpretation.
By the integration of laboratory strategies with automated bioinformatics into a singular multi-analyte test, the scientists can permit patient samples and testing information to remain inside the local laboratory ecosystem and expedite the local lab utilization, subsequently improving far-reaching clinical adoption.
Furthermore, by offering a standardized investigation solution that has received regulatory clearance with a reasonable reimbursement strategy across labs, the interoperability and application of patient test outcomes would be eased, thereby positively influencing the capability to standardize treatment plans from comprehensive genomic profiling tests results.
In this study, the scientists explain the development and analytical validation of the PGDx elio tissue complete test, which contains a 2.2 Mb targeted gene panel, a kitted sample preparation framework, and an accompanying automated bioinformatics examination platform to empower comprehensive genomic profiling of sequence and structural variants, as well as genomic signatures like TMB and MSI in patients with solid tumors.
The Endpoint
The association between mutations in the genome and neoplastic transformation has been deep-rooted, and treatments focusing on tumor explicit genetic abnormalities, both enormous structural changes and small activating sequence mutations, have demonstrated effectiveness across a scope of cancer types.
The identification of patients with the targeted genetic markers is fundamental for successful treatment, and, by and large, single-analyte diagnostic tests have been instrumental in coordinating patient care with targeted therapies.
While NGS targeted panels may not generally be important to evaluate a few significant or prognostic targets utilized for therapy determination in specific clinical settings, they do consider more comprehensive testing of all current and potential future biomarkers from singular sample preparation.
Because of correlations with orthogonal information from two FDA-cleared NGS-based tumor profiling assays, elio tissue complete has shown high analytical performance for the two variants related to therapies or clinical decisions with a PPA of 97.2% and NPA of greater than 99% and variants with potential biomarker importance with a PPA of 85%. These variants can be recognized to biologically relevant levels of 3-6%, below the expected allele fraction for a clonal variant in many tumors.
Even though MSI has been a successful biomarker in several indications, for example, colorectal and gastric cancers, different indications showed relatively low rates of MSI33.
As of late, immune checkpoint blockade has been validated for the treatment of adult and pediatric patients with unresectable or metastatic TMB-high solid tumors that have advanced following prior treatment and who have no good alternative treatment choices.
Going about as a surrogate for neoantigen load, TMB has been displayed to predict response to immune checkpoint blockade. In several studies, the effect of NGS panel size in calculating TMB is investigated.
Smaller panels have been demonstrated to be inaccurate in the evaluation of TMB, with exactness and precision expanding with bigger panels. The latest information proposes that a strong correlation to WES TMB requires a panel of at least 1 Mb, however, increasing panel size to more noteworthy than 1.5 Mb presents little added benefit.
Through the validation of the scientists’ targeted approach, they confirmed high analytical concordance between the predicted eTMB and the entire exome TMB for 307 pan-solid tumor samples.
The high analytical performance of elio tissue complete across an expansive scope of tumor types recommends that this approach would help assess the clinical utility of proposed TMB thresholds (e.g., ten mutations/Mb) in future clinical preliminaries.
A critical boundary to clinical adoption of NGS examinations is the absence of accessibility to validated platforms in local labs. Most approved strategies are presented as send-out services, fundamentally expanding the turn-around time for outcomes and may not be a possibility for all clinicians, for example, those based internationally.
Furthermore, contrasts in sequencing instruments and standardization for targeted panels can bring about variation in TMB determination, making objective assessment problematic.
The scientists have exhibited through these analytical examinations that elio tissue complete is exceptionally unambiguous, precise, and reproducible for the estimation of SNVs, indels, amplifications, translocations, TMB, and MSI.
The combination of the high-performance bioinformatics investigations and a kitted approach to sample preparation with elio tissue complete consider a standardized assessment of biomarkers utilizing a comprehensive genomic profiling test.
This potential for exceptionally precise, standardized outcomes in a decentralized testing environment can empower more cancer patients to approach this comprehensive broad-range testing for their tumor’s particular genetic mutations, facilitating patient access to more successful precision oncology treatment techniques.
Article Source: Keefer, L.A., White, J.R., Wood, D.E. et al. Automated next-generation profiling of genomic alterations in human cancers. Nat Commun 13, 2830 (2022). https://doi.org/10.1038/s41467-022-30380-x
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Tanveen Kaur is a consulting intern at CBIRT, currently, she's pursuing post-graduation in Biotechnology from Shoolini University, Himachal Pradesh. Her interests primarily lay in researching the new advancements in the world of biotechnology and bioinformatics, having a dream of being one of the best researchers.












Excellent