Improved computational tools and artificial intelligence significantly improve drug discovery. This process has automated activities like molecular docking and predictive modeling to discover candidates for drugs with enhanced efficiency and a better prospect of correct predictions. Thus, several cheminformatics tools are currently untouchable for those without relevant specialized expertise. What if there was a way to get students, teachers, and the common man onboard, too? Researchers from the Jagiellonian University and the Polish Academy of Sciences created MedChem Game, which fills the gap by providing a gamified approach to approaching drug design for professionals and novices alike in small-molecule drug development. This is a small mobile app developed for Android that uses the principles of artificial intelligence combined with user-derived data to create a unique learning and discovery interface for drug design.
Need for Accessible Drug Design Tools
Drug discovery is pretty costly and time-consuming and takes years of research and development before a candidate molecule reaches the market. It sometimes requires over $1.3 billion to discover a new drug! To cut the associated costs, the researchers have resorted to computer-aided drug design, CADD for short. CADD uses theoretical and computational methods in modeling and predicting drug-target interactions. It eliminates, to a very large extent, the experimental laboratory trial-and-error. There are generally two major approaches in CADD: ligand-based approaches and structure-based approaches. The ligand-based approaches involve known properties of molecules, but structure-based methods, such as molecular docking, are those that use the 3D atomic structure of target proteins to predict the binding affinities of drug candidates.
These methods are always fairly expert-intensive in chemistry and software use, thereby confining them to students and specialists. That is why the drug design tools developers aimed at democratizing access to these tools- they put them together in a user-friendly mobile game.
What is the Concept of the MedChem Game?
With the MedChem game, users can learn about medicinal chemistry and get involved in discovering drugs by designing small molecules and assessing their potential to bind to biological targets. The gamification elements, points, leaderboards, and a simplified interface are incorporated into the game so that novice and expert medicinal chemists alike can have fun while playing. This must be a dream come true for a chemist who loves gaming! Players can also invent novel drug candidates by sketching molecular structures on an intuitive interface, which are then docked to one of four available protein targets via a remote server. The game provides additional support for a 3D viewer through which the docked complex may be viewed and inspected, as well as the interaction analyzed.
The heart of MedChem Game is a continuously learned deep generative model that learns from user-generated compounds to improve the quality of the newly designed drug candidates. This is a feedback loop because the application not only serves as an educational tool but also contributes to advancement in the field of computer-aided drug design by integrating human creativity into its machine-learning models.
Features of MedChem Game
The features of the application are both educational and entertaining. Users first get to choose their level based on their level of schooling. They are then presented with four protein targets, including monoamine oxidase A and beta-secretase 1, which are associated with conditions like depression and Alzheimer’s disease, respectively. After choosing a target, the user designs their drug candidate using a simple molecular drawing tool that supports drawing by key elements such as carbon, oxygen, nitrogen, and sulfur. This tool simplified molecular design, with the implicit addition of hydrogen atoms and easy modification of bonds, rings, and types of atoms, and, therefore, has opened this game up to all users, regardless of their background in chemistry.
Once a molecule is designed, it is transmitted to a remote server where it docks on the selected protein target using the open-source docking program SMINA, which is AutoDock Vina-based. The docking process predicts the energy when a molecule binds to a protein and provides a score based on the binding affinity. In addition to the docking scores, the game further calculates the Quantitative Estimates of Drug-likeness (QED) and synthetic accessibility (SA) to provide additional information regarding the quality and feasibility of the proposed drug.
There are also training materials, such as onboarding guides, which teach users to use the app along with teaching them the principles behind drug design and molecular docking. So, anyone with absolutely no experience in medicinal chemistry can get up to speed quite easily and begin designing molecules.
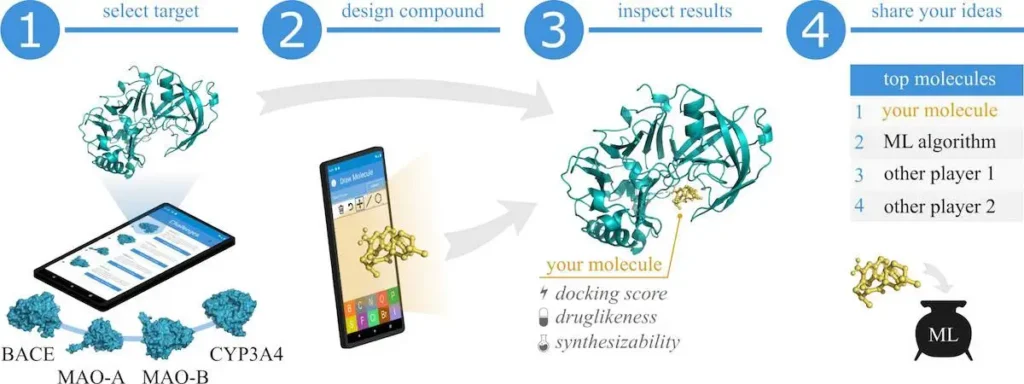
Image Source: https://doi.org/10.1021/acs.jchemed.4c00253
Educational and Research Potential
Similar to Minecraft being used for educational purposes, the MedChem Game can also be used as a tool for teaching students and for medicinal chemists’ research. It has been shown to be an excellent method for teaching principles of drug design, molecular docking, and cheminformatics in the classroom. In lab experiments carried out in high school and university classes, they demonstrated that the game is an effective tool for holding and motivating students. In a student survey conducted after the game-based lessons, 97.5% of students said that they wanted to know more about drug design, and 87.5% said that lectures using the game were more interesting than lectures based on traditional lecturing.
The MedChem Game offers the prospect of an extremely large set of user-designed molecules for researchers. This dataset would be rich in the detail of the design process and the docking scores, thus enabling machine learning models to predict drug-likeness and even binding affinity.
Limitations and Future Directions
Despite the MedChem Game being a major step forward in offering wider open-access access to drug design, it has by no means been free from various limitations. The current game version is unsuitable for professional research projects due to the constraints concerning privacy and other features like ligand-protein interaction analysis capabilities. The drawing tool, although very user-friendly, does not natively support rendering more complicated structures.
To overcome the limitations outlined above, the developers plan to release a research version of the application that will offer greater flexibility in designing molecules, more advanced visualization tools, and better security of the users’ data. Additionally, the developers plan to introduce gamification elements to engage users on a wider scope and multiply the educational effects of the game.
Conclusion
The MedChem Game represents a completely new and exciting platform for education, entertainment, and innovation in drug design. It gamifies the complex process of molecular docking and drug discovery, thus placing these very powerful tools at the doorstep of a different, quite diverse audience-from a student to a professional chemist. Not only is the application teaching medicinal chemistry to a user, but it also inputs toward the field: all the data generated by users can be used to train machine learning models. Additionally, further development could make the MedChem Game not only an educational tool but a strong research tool as well.
Article Source: Reference Paper |The “MedChem Game” application is available for free download on the Google Play Store. The app can be downloaded from the following URL: https://medchemgame.matinf.uj.edu.pl/.
Follow Us!
Learn More:
Neermita Bhattacharya is a consulting Scientific Content Writing Intern at CBIRT. She is pursuing B.Tech in computer science from IIT Jodhpur. She has a niche interest in the amalgamation of biological concepts and computer science and wishes to pursue higher studies in related fields. She has quite a bunch of hobbies- swimming, dancing ballet, playing the violin, guitar, ukulele, singing, drawing and painting, reading novels, playing indie videogames and writing short stories. She is excited to delve deeper into the fields of bioinformatics, genetics and computational biology and possibly help the world through research!
















