Scientists at Kyoto University introduced two new approaches that could help better distinguish RNA modifications affecting how genetic code is read using sequencing technology.
The researchers are getting closer to identifying RNA sequence changes that affect protein formation and can cause diseases. The method, which was published in the journal Genomics, combines probability algorithms with a pocket-sized sequencing device that is already available.
RNA modifications can lead to changes in its structure, function, and stability and play critical roles in gene expression and regulation. According to the authors, all types of biological RNA modifications influence gene regulation, which ultimately determines how different cells function in our bodies.
Anomalies in RNA modifications can result in serious diseases such as diabetes, neurodevelopmental disorders, and cancer. Knowing how and where these RNA modifications occur is critical from a clinical standpoint.
There are already methods for identifying RNA modifications, but those methods are inadequate. For example, with the techniques like chromatography and mass spectrometry, only a small amount of RNA can be processed at a time. On the other hand, high-throughput sequencing methods, which can process large amounts of RNA, necessitate time-consuming sample preparation, cannot map multiple modifications at the same time and are prone to errors. concerning
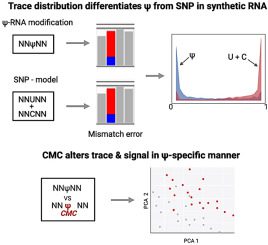
In this study, the researchers present an internal comparison strategy called “IndoC,” in which features like “trace” and “current signal intensity” of potentially modified sites are compared to similar sequence contexts on the same RNA molecule within the sample, eliminating the need for matched knockout controls.
The researchers investigated and discovered two methods for distinguishing a well-known and common RNA modification that involves the substitution of the nucleotide base uracil with another known as pseudouridine.
RNA, like DNA, is made up of strands of varying combinations of four nucleotide bases: uracil, cytosine, adenine, and guanine. The arrangement of these bases determines the code that signals which protein will be produced. When pseudouridine substitutes uracil in the RNA backbone, it can increase protein production or change the code from one that signals information translation interruption to one that signals amino acid formation.
The team’s approach entails utilizing an existing direct RNA sequencing platform developed by Oxford Nanopore Technologies. On this platform, RNA strands pass through tiny pores in a membrane. Depending on the arrangement of the different RNA bases, the current moving through the membrane is disrupted. This lets scientists read the sequence. However, using this approach, scientists frequently struggle to distinguish between different types of modifications.
The researchers developed algorithms to identify a high probability of a pseudouridine substitution versus the possibility of another type of base change.
One of the methods developed by the researchers compares short RNA runs of five nucleotide bases in which uracil, pseudouridine, or cytosine are flanked by the same bases on either side. The readings are then processed by algorithms that determine the likelihood that the middle base is one of the three. The researchers tested their Indo-Compare (Indo-C) strategy on engineered RNA sequences, then on yeast and human RNA, and discovered that it was effective at distinguishing the pseudouridine substitutions from the others.
The authors also detected pseudouridine substitutions by combining a chemical probe with RNA samples and selectively attaching them to them. This altered the sequence readings in such a way that the modification could be identified.
The researchers assume that this work will make nanopore sequencing-based methods for detecting RNA modifications less laborious and more capable of characterizing the effects of these modifications on development and disease.
Further, the research team is looking forward to optimizing the use of both approaches in tandem to identify RNA and DNA modifications more accurately. This will entail the development of new chemical probes that correspond to specific changes. The researchers also intend to continue developing advanced machine learning algorithms to supplement chemical probe-based direct RNA sequencing methods.
Article Sources: Reference Paper | Reference Article
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.