Recent advances in chemistry, material science, and biological research have been greatly aided by large language models (LLMs), which act as flexible foundation models for various downstream predictive or generative tasks. A team of researchers from Zhejiang University and with their collaborators in China proposed an LLM-based foundation model technique called AMP-Designer as a quick way to generate novel antimicrobial peptides (AMPs) with a variety of desirable features. An array of AMPs have been validated in silico tests using AMPDesigner. Results indicate that, with a 94.4% positive rate, the majority of the suggested candidates exhibit noteworthy antibacterial activity. Two candidates in human plasma demonstrated remarkable efficacy, low hemotoxicity, and stability. Forty-eight days were needed to finish the full process, which included in vitro and in vivo validation in addition to in silico design.
Introduction
A significant danger to world health, bacterial antimicrobial resistance (AMR) kills millions of people each year. 1.27 million of the 4.95 million deaths connected to AMR in 2019 were caused by Gram-negative bacteria. By 2050, it is expected that this problem will result in 10 million fatalities annually. The situation is made worse by the absence of novel medications that are effective against Gram-negative bacteria that have grown resistant to widely used antibiotics.
What are Antimicrobial Peptides?
Antimicrobial peptides (AMPs) are naturally occurring proteins that are typically between 10 and 50 amino acids long and are produced by a variety of organisms to fight off invasive invaders. AMPs have been proposed as viable substitutes for traditional small-molecule antibiotics because of their exceptional diversity in structures and functions, outstanding efficacy, and little risk of resistance development.
However, many AMPs have significant drawbacks that prevent their widespread use, such as comparatively lesser antibacterial effectiveness, unclear toxicity profiles, and susceptibility to inactivation during manufacture and transportation when compared to small-molecule-based antibiotics. Current research focuses on creating novel AMP candidates with better resistance to hydrolysis, decreased toxicity, and improved activity to solve these issues. With these enhancements, AMPs ought to function as competing antibiotics. The use of computer-aided technologies to create antimicrobial peptides has gained popularity in recent years. Traditional techniques entail refining already-existing AMPs or screening a sizable peptide space with AMP prediction models. However, finding novel AMPs is difficult due to the large peptide sequence space, which is thought to include 4.5–1041, a peptide with lengths up to 32 residues. There is a need for innovative methods to create AMPs that are more effective because just around ten of them have received regulatory agency approval.
Evolution of AMP Designs with AI
As deep generative modeling techniques have advanced, so have AMP training methods. However, the reliance on supervised learning with limited labeled data makes it challenging to generate AMPs with desired features like reduced hemolysis and high activity. Low novelty is the result of created AMP sequences. A method of sampling from a larger peptide space is suggested to increase novelty; however, this does not provide a high likelihood of producing AMPs with desired characteristics. As alternatives, conditional GANs and conditional VAEs have been developed; however, because they rely on labeled data for training, biases may be introduced, and prediction mistakes may arise. While fine-tuning on small datasets of AMPs with experimental labels is another viable strategy, traditional methods are prone to overfitting and incur significant computing costs.
Understanding AMP-Designer
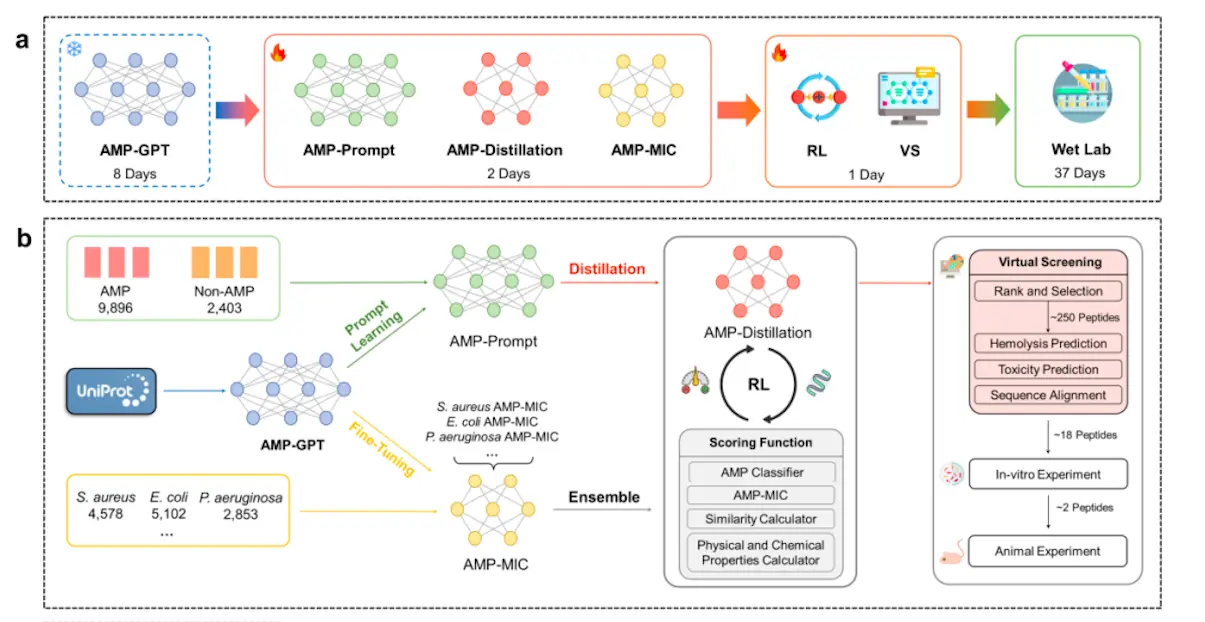
In this work, researchers developed AMP-Designer, a comprehensive framework for designing artificial general-purpose machines (AMPs) that includes GPT, prompt tuning, contrastive learning, knowledge distillation, and reinforcement learning (RL). A series of wet-lab evaluations are then conducted to validate the newly designed AMPs. Using contrastive prompt learning, the model is trained on a dataset of peptides that were taken from UniProt in order to help in transfer learning on a labeled dataset. AMP-Prompt is subjected to model distillation, which results in an RNN made up of three GRU layers. For various bacterial species, the minimum inhibitory concentration prediction models, or AMPMIC, are built using AMP-GPT and offer feedback while easing RL screening. This versatile technique is a breakthrough in biomedical research, providing significant advantages over current models in both computational and experimental contexts.
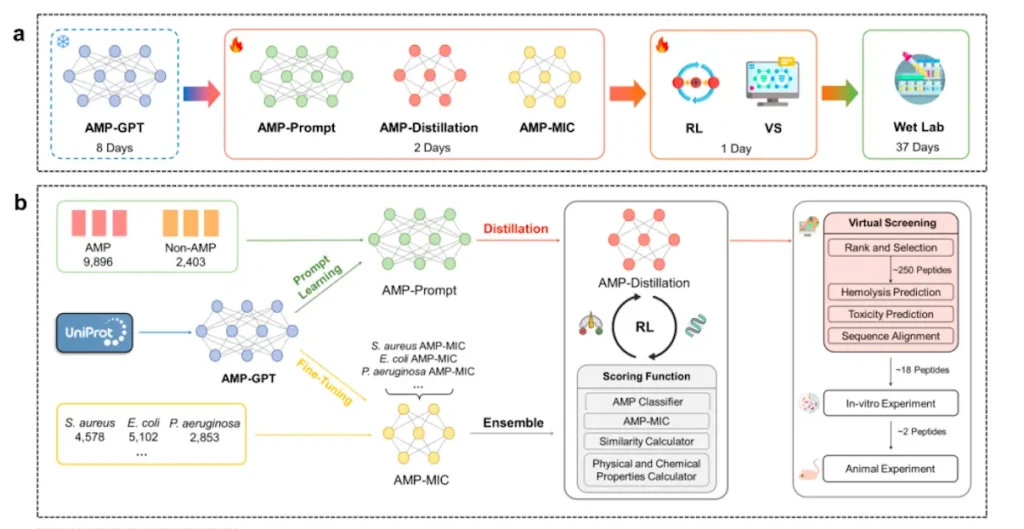
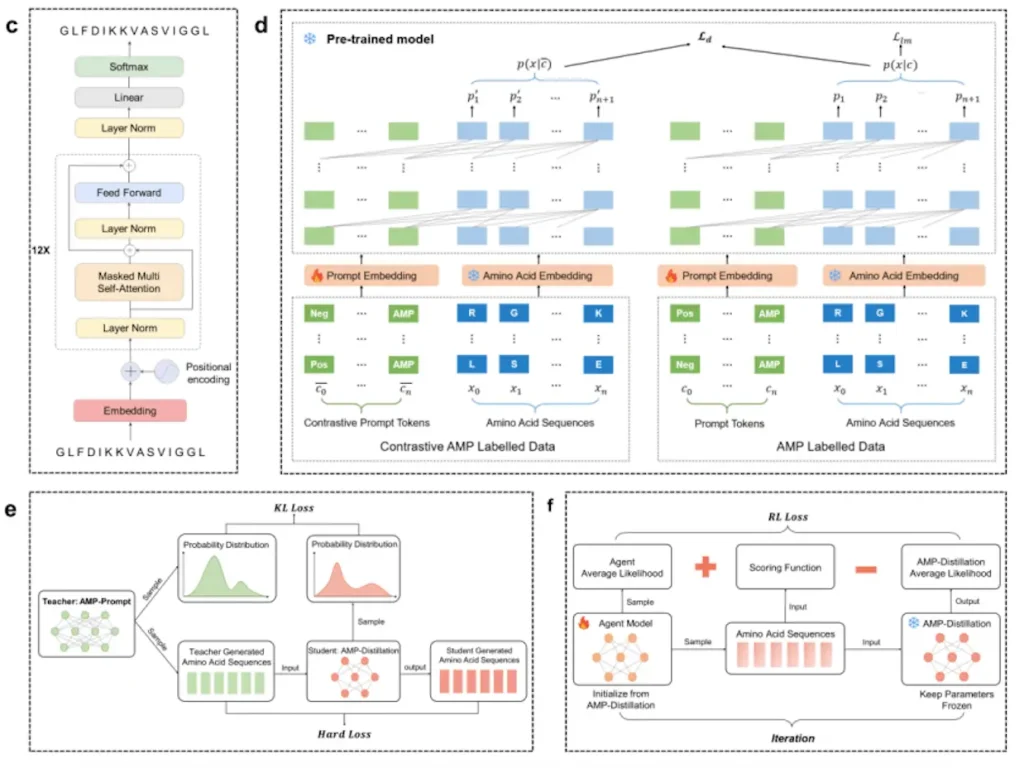
Applications of AMP-Designer
- Potent AMPs were created using AMP-Designer to combat various Gram-positive and Gram-negative bacteria. In less than 48 days, two remarkable AMPs were identified: KW13 and AI18. The strong in vitro antibacterial activity of these peptides was shown to be effective against both Gram-positive and Gram-negative bacteria. Against therapeutically generated resistant Gram-negative bacteria, they showed high antibacterial activity, good plasm stability, and little off-target hemolysis damage. Additionally, KW13 and AI18 demonstrated exceptional therapeutic efficacy in a mouse model of bacterial pneumonia.
- AMP-Designer is a robust framework that leverages the trained foundation model AMP-GPT to expeditiously accomplish design tasks. Particular AMP design is one of its strong points, even in situations with little labeled data. For instance, Propionibacterium acnes, an anaerobic bacterium with few labeled data points, was the target of AMPs designed using this technique. Three of the five AMP candidates that the framework produced showed great potency in evaluations conducted through experiments. The efficiency of the AMP foundation language paradigm in the antimicrobial peptide discovery process is demonstrated by this successful few-shot AMP design.
Conclusion
A novel framework for designing AMPs, the AMP-Designer model combines reinforcement learning, knowledge distillation, and quick tweaking. With an amazing activity prediction rate of 83.4% for AMPs employing three SOTA AMP categorization predictors, this model greatly improves the efficiency of AMP generation. Reinforcement learning is another technique the framework uses to optimize the model for better activity. The model was used to generate antimicrobial peptides against Gram-negative bacteria in order to validate its performance; 18 peptides were chosen for experimental validation.
Article Source: Reference Paper | The code used in the study is publicly available on the GitHub repository.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.