Researchers from MIT have created a comprehensive map of over 300 protein kinases and their targets, with the goal of potentially discovering new cancer treatments.
Protein kinases are a vital class of enzymes that play a crucial role in the human body. These signaling molecules control a wide range of cellular activities, including growth, cell division, and metabolism. When these pathways malfunction, it can result in various diseases, with cancer being the most prevalent.
Protein Phosphorylation
Protein phosphorylation is a common post-translational modification in biology. Recent advances in mass-spectrometry-based phosphoproteomics have allowed scientists to study this process in more detail. So far, they have identified 90,000 sites on proteins where this happens and have found that many of these are linked to disease or other important cell processes. However, scientists still need to find out which of the over 300 enzymes in the human genome are responsible for most of these phosphorylation events.
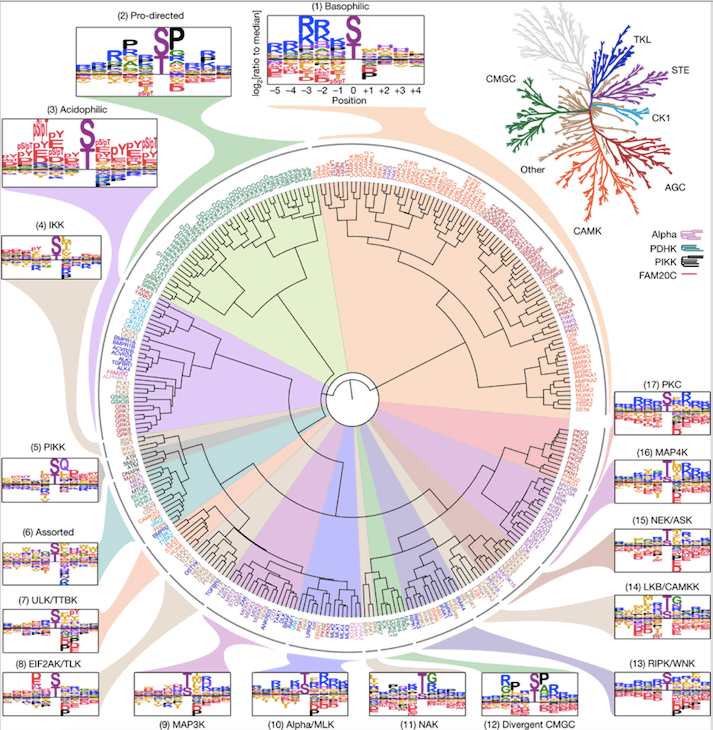
Human Serine/Threonine Kinome
Uncovering the protein kinases that contribute to cellular malfunction and cancer progression has the potential to reveal a plethora of new drug targets. However, for the majority of these kinases, researchers need a comprehensive understanding of the pathways in which they play a role or the specific molecules on which they act.
Despite the abundance of cancer genome sequencing data, a comprehensive examination of signaling pathways and protein kinase activation states in cancer is lacking. With this information now available, a more targeted approach to treating tumors can be developed.
The researchers recently developed a comprehensive atlas featuring over 300 human cell protein kinases, uncovering their probable targets and regulation. This breakthrough could assist researchers in unraveling numerous cellular signaling pathways and aid in understanding how these pathways are affected when cells transform into cancer or are exposed to particular medications.
The human genome comprises a vast array of protein kinases, numbering over 500, that regulate other proteins through a chemical process called phosphorylation. Despite their significance, the specific proteins targeted by most of these kinases remain uncharted. However, extensive research on kinases such as MEK and RAF, which play a vital role in the control of cell growth, has led to the development of innovative cancer medications that block these kinases.
Scientists have long used mass spectrometry to uncover proteins that are differently phosphorylated in cancer cells compared to healthy cells to identify unique pathways that are disrupted in cancer. However, the challenge with this method is that it’s hard to determine which enzyme kinases are responsible for phosphorylating these proteins, thus leaving the regulation and disruption of these proteins in disease as a mystery. With the new technology, researchers can now easily analyze mass spectrometry data and reveal which protein kinases are accountable for phosphorylating the proteins, providing more insight into the regulation and disruption of these proteins in disease.
While we have a wealth of data on phosphopeptides, it remains to be determined how they fit into the larger picture of cellular signaling pathways. With a key to decode this information, it is easier to determine which pathways are being affected by these peptides. The root of this problem is a lack of knowledge about the specific substrates for most protein kinases.
In this research, the team examined the substrate specificities of two major categories of kinases, i.e., serine kinases and threonine kinases, which constitute a significant 85% of the human body’s protein kinases, by analyzing the structural patterns onto which they transfer phosphate groups.
The researchers employed a library of peptides created previously to discover patterns that kinases interact with. They gauged the interactions of the peptides with all 303 known serine and threonine kinases. They then employed a computational model to study the interactions and were able to recognize the kinases that can phosphorylate all 90,000 known phosphorylation sites in human cells for both classes of kinases.
The scientists discovered that many kinases, despite having dissimilar amino acid sequences, have developed the ability to bind and catalyze phosphorylation on similar substrate motifs. Further analysis revealed that approximately half of the kinases explicitly examined targeted one of three principal classes of motifs, while the other half specialized in phosphorylating one of roughly a dozen subclasses of motifs.
Atlas of Kinase Substrate Specificities
The kinase atlas substrate specificities can assist scientists in recognizing dissimilarities in signaling pathways between normal and malignant cells or between treated and untreated cancer cells.
The newly developed atlas of kinase motifs has proven to be a valuable tool in decoding the intricacies of signaling networks. Studying phosphorylated peptides can identify which specific kinase is responsible for each reaction, thus giving a clearer understanding of the entire process.
The researchers tested their approach by examining cells exposed to an anti-cancer medication targeting Plk1. This specific kinase plays a key role in cell division. As anticipated, the analysis of phosphorylated proteins revealed that a significant number of them were controlled by Plk1. However, the study also revealed that the treatment had an unexpected side effect, leading to a boost in the activity of two other kinases that are both involved in the cell’s response to DNA damage.
The researchers are now looking forward to utilizing the enzyme atlas to identify other abnormal communication pathways that cause cancer, particularly in certain cancer types that lack genetic causes discovered yet.
Phosphoproteomics is a new way to study cancer. It allows us to see which pathways in the cancer cells are upregulated or downregulated. This can help figure out what’s causing cancer, even when it’s not clear what the genetic cause is.
Conclusion
The development of an atlas of substrate specificities for the human serine/threonine kinome represents a significant advancement in our understanding of cellular pathways. By providing a comprehensive map of the interactions between enzymes within the cell, this atlas allows researchers to gain new insights into the complex network of reactions occurring within cells. This knowledge can be used to develop new drugs and therapies that target specific enzymes and pathways, potentially leading to more effective treatments for diseases such as cancer.
Article Source: Reference Paper | Reference Article
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.