For many years, scientists have been baffled by the difficult task of designing enzymes, which is regarded as one of the most difficult challenges in biochemistry. However, the introduction of new techniques that allow the creation of scaffolds tailored to specific functions has provided new hope. The research published in Nature reports the production of highly active and specific biocatalysts from scratch, with broad applications in biomedicine and various biotechnological applications. The approach enables the generation of a wide range of luciferases and other enzymes.
The researchers report the development of extremely effective enzymes using computational methods that differ from any naturally occurring ones. It has been demonstrated in laboratory experiments that these newly developed light-producing luciferases can successfully identify and catalyze the emission of photons from specific chemical substrates in a highly efficient manner. This advancement in protein design is significant because enzymes are widely used in a variety of fields, including biotechnology, medicine, environmental cleanup, and manufacturing.
The team started developing novel luciferases by handpicking the luciferins they wanted the proteins to act upon. They then used software to generate many potential protein structures that could interact with those luciferins. The best-performing enzymes generate enough light intensity to be visible without the use of any visual aids.
Creating protein folds
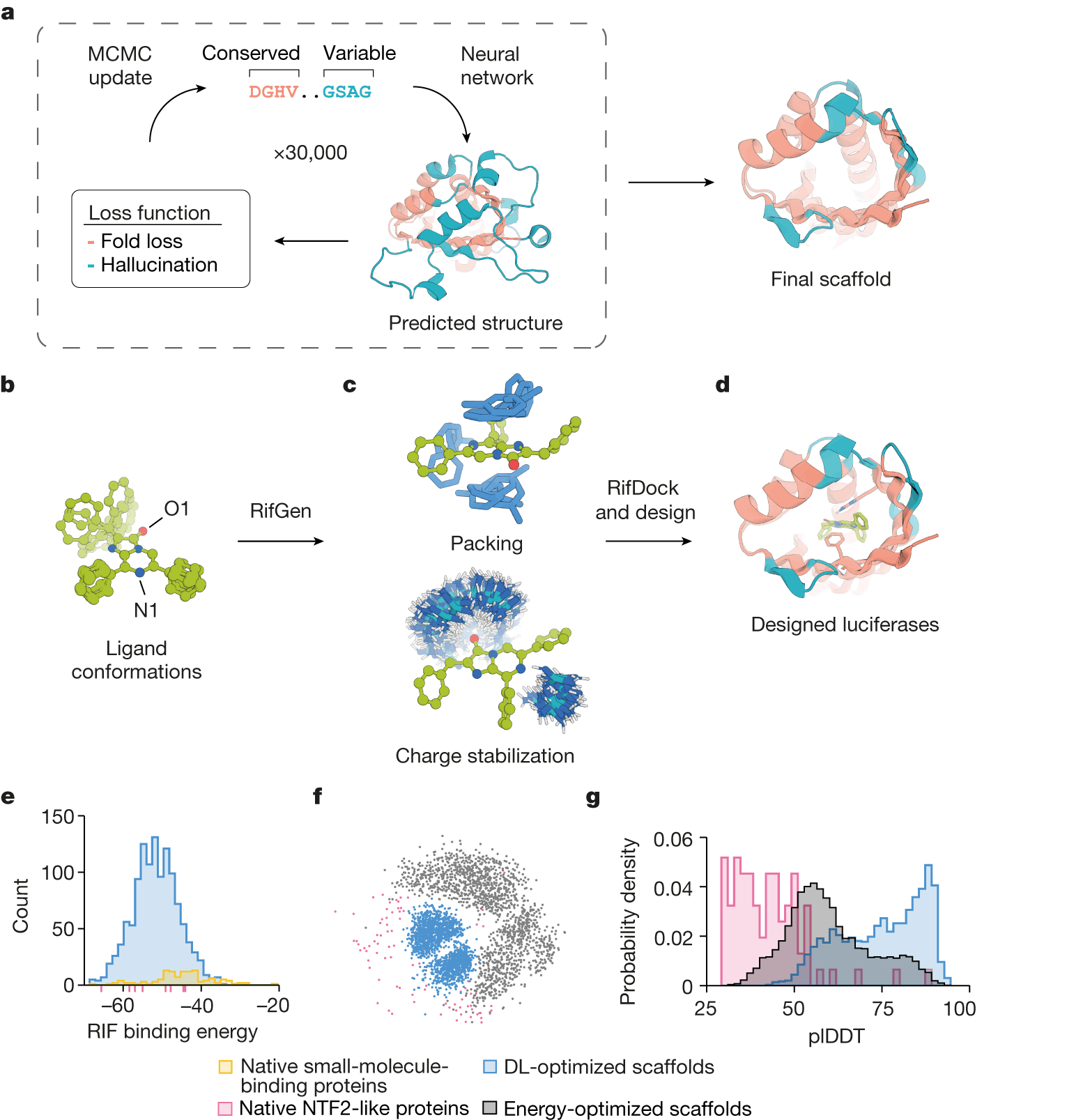
Instead of modifying existing proteins, the researchers developed a novel protein design technique called ‘family-wide hallucination’ using deep learning. This method combines unconstrained de novo design with fixed backbone sequence design to generate an infinite number of previously unseen proteins with a desired fold.
The hallucination of proteins across the entire family entails using unconstrained protein hallucination to discover new sequences and structures for loop and variable regions while using structure-guided sequence optimization for core regions.
To incorporate active sites into the imagined protein structures, the researchers chose to precisely position a positively charged guanidinium group from an arginine residue in order to stabilize the negative charges that exist in the reaction’s transition state. Additionally, more active site residues were also designed.
Let the light shine
The scientists discovered three functional enzymes from their initial models after producing and testing them in the laboratory. The most effective one was designated LuxSit, which was a clever reference to the Latin phrase of UW’s motto, lux sit, meaning ‘let light exist.’
LuxSit has a number of characteristics that make it an appealing tool for biotechnological research. It is the smallest luciferase known, with only 117 residues. When the enzyme was incubated with its synthetic luciferin substrate diphenylterazine (DTZ), it produced blue luminescence at 480 nanometers, which is consistent with the chemiluminescence spectrum of the substrate. Under near-boiling conditions, the protein was found to remain partially folded.
LuxSit’s enhancement resulted in a significant improvement in its operation. LuxSit-i, a refined enzyme, produced enough light to be seen without assistance. It was discovered to be more luminous than the luciferase enzyme found naturally in Renilla reniformis, a luminous sea creature.
Rather than relying on naturally occurring enzymes, the scientists were able to create highly effective enzymes using computer-based design. This breakthrough suggests that personalized enzymes for nearly any chemical process may be possible.
Deep learning for protein design
The researchers then developed additional luciferases capable of detecting a different artificial luciferin compound, 2-deoxycoelenterazine or h-CTZ.
Because h-CTZ and DTZ have different molecular structures, the researchers used a family-wide hallucination technique to create personalized protein scaffolds. Based on the most effective models observed during the initial luciferase design phase, they introduced specific configurations of histidine and arginine side chains to create active sites.
Instead of RosettaDesign, the team used the newly developed ProteinMPNN tool to finalize the amino acid sequences of the new enzymes. This sequence design tool is powered by deep learning and has been shown to outperform previous software. Additionally, it operates in less than one second, surpassing its predecessors by more than 200 times. The best part is that it can function properly without any professional modifications.
Two of the 46 h-CTZ catalysts created and analyzed in the laboratory exhibited luciferase activity that could be measured. Compared to the previous round of de novo enzyme design, which had a success rate of only 0.04%, this represents a significant improvement in success rates (3 out of 7,648 for DTZ). The improved success rate of 4.35% for h-CTZ in this second round is most likely due to the valuable knowledge gained during the first design round and ProteinMPNN’s improved performance.
Conclusion
Progress in developing enzymes via computational methods was hampered by a scarcity of protein frameworks and the difficult task of embedding active enzyme sites within them. However, advanced techniques such as deep learning to create many personalized protein frameworks and improved tools for protein sequence design have paved the way for a new phase in enzyme development.
Article Source: Reference Paper | Reference Article
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.