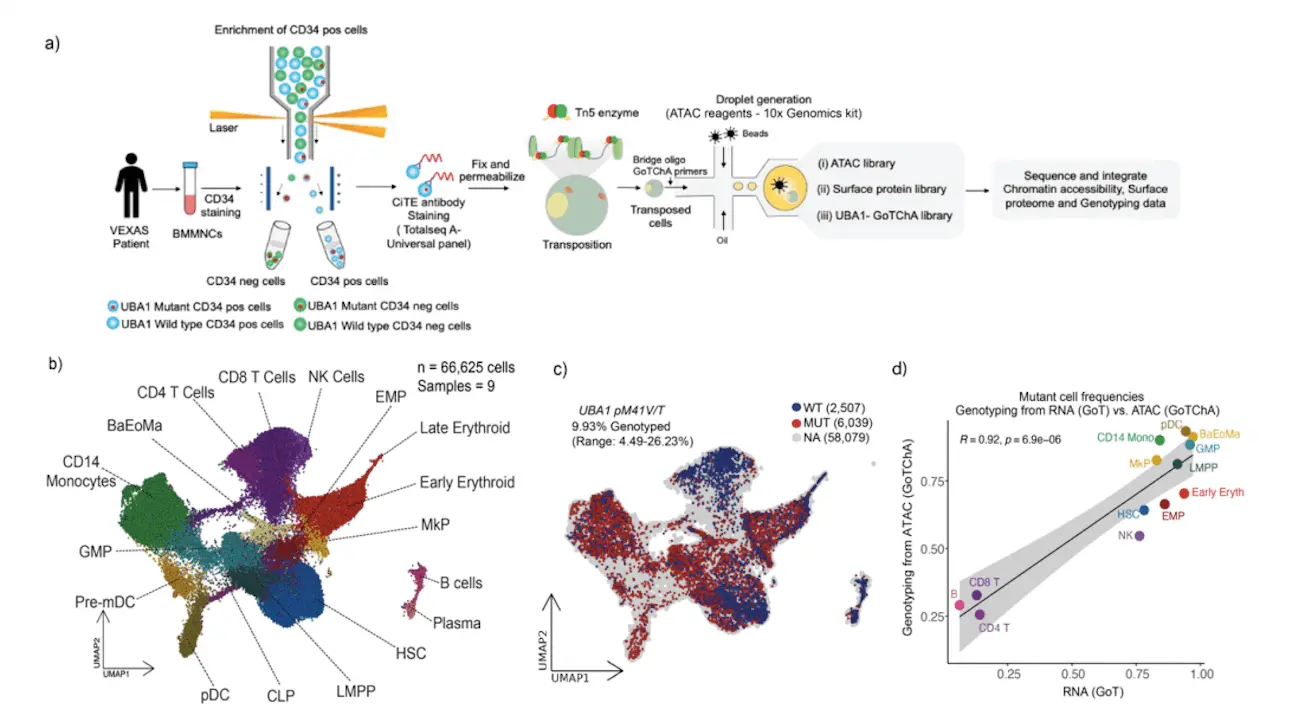
Clonal variation emerges across tissues as a result of somatic evolution, with far-reaching consequences for human health. The VEXAS (Vacuoles E1 enzyme X-linked Autoinflammatory Somatic) disease, which is brought on by somatic UBA1 mutations in hematopoietic stem cells (HSCs) and results in treatment-refractory systemic inflammation, is a remarkable illustration of somatic evolution. However, the lack of knowledge about the mechanisms behind the survival and proliferation of mutant HSCs hinders the creation of efficient treatments.
Such a mechanistic understanding has been impeded by the paucity of animal or cellular models of UBA1-mutant HSCs, necessitating the investigation of primary human VEXAS samples that contain combinations of wild-type and UBA1-mutant HSCs. In order to overcome these obstacles, scientists used single-cell multi-omics to thoroughly characterize the chromatin accessibility, signaling pathway alterations, and mutant UBA1-induced transcriptome in VEXAS patients. This allowed for the direct comparison of mutant and wild-type cells in the same setting. New understandings of disease characteristics were provided by the anticipated enrichment of UBA1M41V/T mutations in myeloid cells and Natural Killer (NK) cells in VEXAS patients. Mutant cells carrying the UBA1M41V/T mutation displayed elevated indicators of inflammation together with the unfolded protein response (UPR) triggered by pro-survival mediators of the PERK pathway. PERK inhibition reversed the effects of up-regulating the PERK pathway, increasing myeloid differentiation and cell survival and inducing UBA1 in normal CD34+ or UBA1-mutant HSCs. This implies that blocking the PERK pathway could be a useful treatment approach to get rid of the clone that causes VEXAS.
Introduction
According to recent discoveries, the human genome is made up of many diverse clones that are driven by somatic driver mutations. Both benign illnesses and normal tissues exhibit these clonal expansions. The most extensively researched area of non-malignant somatic evolution has been the hematopoietic system, which has demonstrated that clonal expansions are almost universal in hematopoietic stem cells (HSCs). By the age of 70, this has resulted in a high prevalence of clonal hematopoiesis of indeterminate potential (CHIP), which has been connected to a number of metabolic, autoimmune, and cardiovascular disorders.
What is VEXAS Syndrome?
The vacuoles E1 enzyme Xlinked autoinflammatory somatic (VEXAS) syndrome is a unique somatic condition caused by somatic mutations in the ubiquitin-activating enzyme E1 gene that starts in human stem cells (HSCs). Misfolded protein accumulation, systemic inflammation, and the unfolded protein response (UPR) are caused by these mutations. Only 63% of patients with VEXAS syndrome survive for five years. The cellular ubiquitination process is initiated by UBA1, but UBA1-mutant cells generate a third isoform (UBA1c) that is unable to correctly ubiquitinate proteins, which causes inflammation and biases development towards the myeloid lineage. Because researchers don’t know much about the survival mechanisms of UBA1-mutated HSCs, current medications focus on reducing inflammation rather than the mutant HSCs that cause the disease. The pathogenic implications of the UBA1 mutation on disease-initiating HSCs, a population of fewer HSCs, are the main subject of the study. The goal of the study is to better understand these HSCs’ phenotypes, which are still poorly understood. Important issues are whether inflammation and myeloid bias originate from HSCs or other progenitor cells, how mutant HSCs expand and survive in the face of high protein toxicity, and whether phenotypic abnormalities caused by somatic UBA1 mutations indicate underlying vulnerabilities of the cell.
The combination of wild-type and mutant cells presents a major obstacle to the investigation of somatic clonal expansions in humans. Through high-throughput screening of both wild-type and mutant cells, single-cell multi-omics approaches enable the characterization of changes to the transcriptome, epigenome, and proteome that are unique to mutant cells. With this method, patient-specific confounders in human studies are eliminated, and it is possible to compare mutant and wild-type cells directly within the same subject. It is able to recognize variations unique to mutants, which may act as therapeutic openings to eradicate mutant cells.
Understanding Single-cell Analysis in VEXAS Syndrome
The study focuses on the pathogenesis of VEXAS and finds parameters that may be addressed to eliminate HSCs that initiate illness. Through the use of multi-omic single-cell methodologies, such as Targeted Loci with Chromatin Accessibility (GoT-ChA) and Genotyping of Transcriptome (GoT), the researchers discovered mutation enrichment in both natural killer (NK) and myeloid cells. The PKR-like endoplasmic reticulum kinase (PERK) branch of the UPR pathway was implicated in the myeloid priming of mutant cells, enhanced inflammatory signals, global translational downregulation, and higher ATF4 target gene enrichment that were found during analysis of HSCs. Inhibition of the PERK pathway lowers ATF4 expression, which is correlated with decreased CD34+ cell survival, according to functional validation of the PERK pathway’s role in HSC fitness. The findings highlight the importance of single-cell genotype-to-phenotype mapping in human somatic mosaicism and imply that targeting the PERK pathway in HSCs may constitute a novel approach for the treatment of VEXAS syndrome.
Inflammatory and Stress Responses Unique to Cell-types Revealed by Single-cell Analysis in VEXAS Syndrome
Cell type affects somatic evolution in VEXAS, and somatic mutations play a major role in this process. Researchers analyzed CD34+ enhanced bone marrow progenitor cells from VEXAS patients and healthy controls using GoT and CITE-seq. After data analysis, it was discovered that VEXAS HSCs had a myeloid skew when compared to healthy controls. Differential gene expression showed depletion for translation pathways and enrichment in pathways related to unfolded protein response, proteome stress, and inflammation. This indicates that the UBA1 mutation mediates the disruption of the protein degradation pathway, underscoring the significance of cell type in VEXAS pathogenesis. The work emphasizes the necessity for more investigation into the pathophysiology of VEXAS and somatic mutations.
Conclusion
The phenotype mapping of VEXAS syndrome using a single-cell genotype revealed fundamental abnormalities in somatically altered cells that support HSC survival. Our findings thus show the potential of single-cell multi-omics analysis of somatic mosaicism in primary patient samples for the identification of new targets for the precise targeting of mutant cells and the identification of disease processes.
Article Source: Reference Paper | All analysis scripts are available on https://github.com/RebeccaMurray/VEXAS-scRNA-seq and https://github.com/RebeccaMurray/VEXAS-scATAC-seq.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.