A thorough understanding of the cellular connections inside these tissues is being made possible by technologies that use spatial transcriptomic approaches to revolutionize the study of ligand-receptor signaling in tissues. CytoSignal was created by researchers from the University of Texas and the University of Michigan to use spatial transcriptomic data to infer the locations and dynamics of cell-cell communication at the cellular level. CytoSignal offers a basic understanding of the spatial dynamics of signaling connections. It finds differentially expressed genes, measures contact-dependent and diffusible interactions, and locates geographic gradients in signaling strength. Numerous spatial transcriptomic approaches, such as spot-based protocols without deconvolution and FISH-based techniques, are compatible with CytoSignal. The tool’s outcomes are verified in situ using a proximity ligation assay, which shows that tissue locations of ligand-receptor protein-protein interactions closely correspond with CytoSignal scores. The current necessity for cellular resolution detection of cell-cell signaling connections and their dynamics is met by this dependable and scalable method.
Introduction
In multicellular animals, cell-cell communication is an essential mechanism that requires the dimerization of membrane-bound proteins or the binding of secreted ligands to transmembrane receptors. Differentiation, destiny selection, immunological response, growth, and physiological tissue function depend on this communication. Finding the signaling relationships between cells in particular situations is still difficult, though. Some progress has been made by elucidating the expression of ligands and receptors by cell type within diverse tissues using single-cell RNA sequencing (scRNA) datasets. Techniques for inferring cell-cell communication from single-cell RNA data have been reported, such as CellPhoneDB, CellChat, NicheNe, SingleCellSignalR, and Scriabin. However, these methods lack information regarding cell spatiality and cannot be used to infer signaling among cell groups.
Gene expression profiles and spatial coordinates inside a tissue are measured using spatial transcriptomic techniques, such as CellPhoneDB v4.0 and SquidPy. This information is vital for determining the spatial closeness of cells producing ligands and receptors. These techniques concentrate on interactions between sizable designated cell groups, just like the majority of scRNA data.
The process of signaling entails interactions between cells in tissue via proteins, with the spatial microenvironment dictating the sites of these contacts. When spatial context is given, traditional Spatial Transcriptomics (ST) approaches based on abstractions are limited because cells do not verify the types of other cells before interacting. The spatial aspect of ST data is not properly utilized by these approaches to infer signaling connections at the cellular level. Furthermore, different kinds of signaling interactions, like those involving diffusible ligands and transmembrane receptors or contact-dependent interactions, are modeled in the same way by current techniques for scRNA and ST data. Moreover, current methodologies just deduce the present condition of cell signaling during the time of cell measurement, failing to account for the evolution of these interactions throughout time.
Understanding CytoSignal
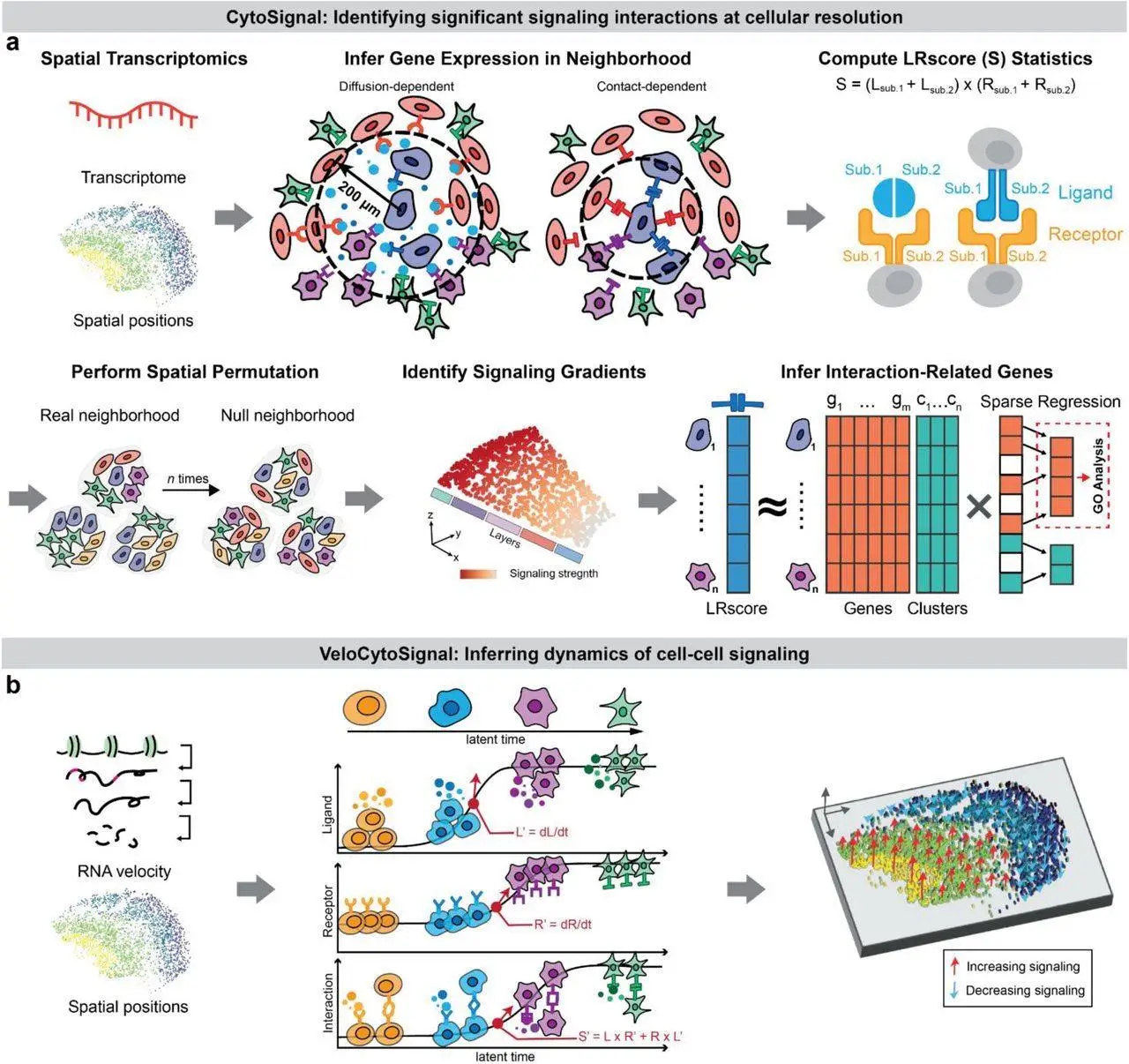
CytoSignal is a unique method for calculating static and dynamic signaling among single cells at precise spatial coordinates by using spatially resolved gene expression. To determine whether cells and regions within a tissue exhibit considerable activity for a specific signaling connection, CytoSignal employs a straightforward, principled score. VeloCytoSignal, a technique for forecasting the rate of change for a signaling interaction at each tissue location, was further refined by researchers.
In cell-cell communication investigations, confirming inferred signaling processes in living tissues is the main hurdle. Previous approaches frequently depend on computational validation, which fails to show the methods’ accuracy and practicality in real-world tissue settings. Predictions made by CytoSignal, a ligand-receptor interaction predictor, can be verified more precisely by creating an in situ validation analysis using the proximity ligation assay (PLA). The physical sites where a ligand binds to its receptor are verified by this study, offering concrete proof in support of CytoSignal’s predictions in actual tissues.
Identify Locations and Dynamics of Signaling Interactions at Cellular Resolution by CytoSignal
The fundamental concept of CytoSignal is that protein-protein interactions at particular tissue locations drive ligand-receptor signaling. When every necessary element is expressed in close spatial proximity, this interaction takes place. Based on the ligand-receptor interaction score, a score is created to indicate the strength of these interactions at every location inside a tissue (LRscore). An underlying chemical process between the ligand and receptor proteins that results in the formation of a complex drives the score. This technique offers a straightforward, comprehensible measure of signaling activity at every location across the tissue. The score enables the quantification of the signal received by every cell and the inference of signal-sending cells for every signal-receiving cell. It may also be used to evaluate contact-dependent versus diffusion-dependent interactions.
By measuring the degree of signaling contacts in tissue, researchers may utilize CytoSignal to pinpoint important cell locations and genes linked to signaling. CytoSignal can detect cells with high signaling activity and spatial gradients in signal strength by flipping the positions of the cells and computing a p-value. Unlike prior approaches that rely on cell type annotations, the methodology presented in this study enables continual change in signaling across a tissue. Furthermore, CytoSignal makes it possible to identify genes that are differentially expressed in relation to a signaling relationship, providing objective search and control over the influence of cell type. GO keywords or transcription factors connected to the signaling interaction can then be found using this information.
Conclusion
CytoSignal is a cell-level method that uses spatial transcriptomic data to infer the dynamics and activities of signaling relationships at each spatial point inside a tissue. Finding genes linked to signaling, identifying contact-dependent vs. diffusion-dependent connections, and identifying spatial gradients in signaling are all made possible by this cell-level method. Numerous tissue contexts, such as the developing mouse embryo, adult mouse brain neurogenesis, and neurodegenerative models, have seen the beneficial application of CytoSignal. It can uncover signaling networks in a variety of physiological and pathological situations and is compatible with the majority of spatial transcriptomic approaches. In addition to revealing interactions between immune cells, healthy cells, and malignant cells in the tumor microenvironment, the technique seeks to increase applicability across different data modalities.
Article source: Reference Paper | CytoSignal and VeloCytoSignal R package is available on GitHub
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.