Since the COVID pandemic began, the world has been reminded of the poor reliability of elevated body temperature for detecting infections. Even when infected with SARS-CoV-2, many patients exhibit no fever. But while obvious symptoms can be absent, infections trigger host gene responses at sites harboring pathogens and in distant cells receiving molecular signals. Leveraging this, researchers now report a saliva test that analyzes human gene activity to identify diverse infections with over 90% accuracy.
The Failed Promise of Fever for Detecting Infection
For over a century, body temperature has served medicine as a crude proxy for identifying acute infections based on the commonly held notion that infectious pathogens trigger fever. However, modern observations have repeatedly affirmed the limited sensitivity of fever, especially early in infections.
During the early phase of the COVID-19 pandemic, temperature checks aiming to identify SARS-CoV-2 infected individuals proved widely ineffective. A substantial proportion of transmission owed to asymptomatic and presymptomatic carriers with no discernible fever. The subsequent realization that infection abundantly spreads without symptoms made clear the inadequacy of fever for diagnosis and surveillance.
In truth, countless preceding studies with influenza, malaria, gastroenteritis, and more also demonstrated poor diagnostic performance of elevated temperature. Even febrile patients exhibit inconsistent fever heights regardless of infection severity. After over 100 years of falsely elevating its clinical utility, the pedestrian thermometer has shown its failure to serve as an accurate molecular indicator of acute infectious threats.
Seeking a Better Molecular Thermometer
While fever provides little consistent signal, infections reliably trigger host molecular responses – even without symptoms. Upon recognizing pathogens, sentinel cells switch on networks of genes to eliminate the threat and coordinate defenses. Classes like interferons and inflammatory cytokines activate cascades of downstream antiviral genes.
Measurements of these gene activation signatures offer alluring molecular proxies for infection with far greater sensitivity than fever peaks. However, prior to the emergence of genomics, directly monitoring infection-induced gene activity in patients remained impractical.
By exploiting modern “omics” assays, scientists now envisaged unlocking signature host responses to pathogens as a novel diagnostic indicator. The vision was effectively developing a molecular thermometer – using human gene expression changes to accurately classify infection regardless of clinical presentation. The ultimate goal was to roll out accessible screens using easily obtained biosamples like saliva or blood.
Finding a Core Host Response to Infection
To pioneer this, a multi-institutional team spearheaded by scientists at Stanford and UCSF first compiled diverse public data on human cells infected with various viral, bacterial, and fungal pathogens in lab experiments. Cross-referencing differential expression analyses of infected vs healthy host cells revealed a conserved set of around 70 human genes consistently upregulated by over 80% of tested disease agents.
Dubbed the “core response” genes, this shared signature demonstrated activation in response to numerous viruses, bacteria, and fungi – rather than a narrow pathogen class. Many resided in expected immune pathways governing interferon stimulation and inflammation. But rather than assuming a select few genes would respond, the researchers empirically derived this infection sensor array from scratch by data-driven integration across pathogens.
Further computational experiments confirmed a consistent signal regardless of particular datasets used – with levels of these 70 genes alone powerfully discriminating infected from healthy cells using straightforward statistical classifiers. This supported the existence of a fundamental host response to pathogenic invasion.
Developing a Saliva Test for Human Trials
With a candidate genomic infection signature identified, the researchers first demonstrated that expression of these core response genes reliably manifests in patient saliva and blood when infected. Intriguingly, elevated RNA levels proved substantially more prominent in saliva – identifying this easily collected bodily fluid as an ideal substrate for rapid diagnostic tests.
Subsequent investigation of the RNA expression levels of core response genes in infected individuals’ saliva samples showed impressive consistency. Across multiple patient cohorts – spanning SARS-CoV-2 to cholera and skin infections by diverse bacteria – salivary RNA abundance of around 70 identified genes enabled sensitive molecular identification of infected patients using simple computational models.
By forgoing complex statistical pipelines in favor of straightforward classifying algorithms, the researchers emphasized the robustness of the host response expression signal in saliva for tangibly separating infected from healthy.
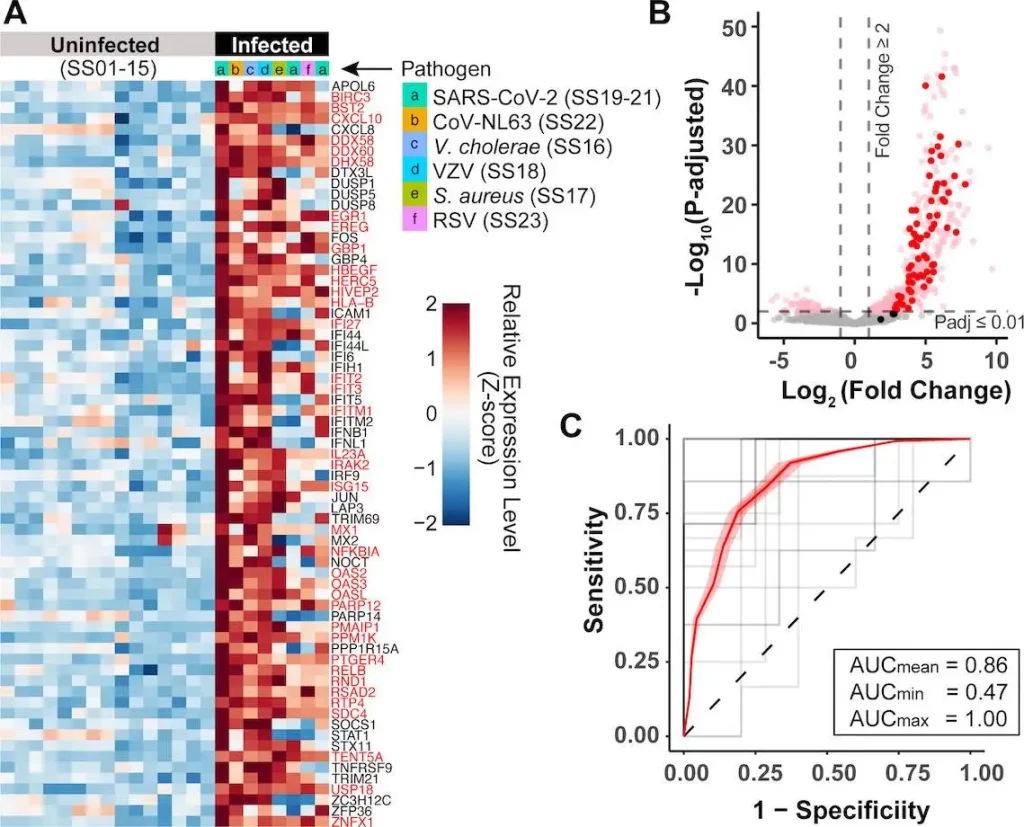
Image Source: https://doi.org/10.1128/mbio.01712-23
Classifying Asymptomatic SARS-CoV-2 Infections
As the most telling demonstration, the team lastly analyzed a cohort of university students and staff undergoing mandatory but asymptomatic SARS-CoV-2 saliva screening. Despite the complete absence of symptoms, host gene expression levels reliably diagnosed COVID-19 infection status with a remarkable 87% accuracy. By comparison, symptom screening entirely failed by design among these infected yet presymptomatic individuals.
Just as importantly, the salivary molecular test effectively flagged carriers with viral titers indicating likely infectiousness for transmission. Capturing these hidden asymptomatic threats by objectively detecting the endogenous host response rather than relying on subjective self-perceived symptoms remains essential for outbreak control amidst pathogens like SARS-CoV-2. The findings illuminate the power of tapping directly into host physiology for infection diagnostics.
Envisioning the Future of Diagnostic Testing
This research provides a glimpse into a future no longer dependent on crude nonspecific indices like fever but instead harnessing molecular sensors revealing the body’s native immune response to identify threats objectively. Already possible today, such technologies could readily support field-deployable rapid saliva screening to guide clinical triage, contact tracing, and surveillance amidst outbreaks.
By moving decisively beyond the thermometer, infection-induced human gene expression biomarkers promise 21st century diagnostics at the molecular level – with the requisite speed, objectivity, and sensitivity for preparedness against perpetually circulated and emergent pathogens.
Conclusion
Of course, scaffolding these experimental findings into widespread point-of-care devices requires confronting myriad scientific, engineering, regulatory, and implementation hurdles. Follow-up studies systematically benchmarking performance across populations, confounding illnesses and stages of infection will prove essential. But the conceptual foundation has been laid for saliva gene expression tests to provide a testament that while infections may frequently evade obvious clinical notice, they cannot escape divulging their presence to the watchful host.
Article Source: Reference Paper
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.















