Personalized Drug Therapy is growing rapidly nowadays. For that, accurate representations of the protein structures are important. Recently, the use of machine learning and deep learning techniques for the unsupervised learning of protein representations has been studied extensively. Still, they often focus only on the amino acid sequence of proteins and lack knowledge about proteins and their interactions, thus impacting their performance. Researchers from Technion and Meta AI present GOProteinGNN, a novel framework that enhances protein language models by combining protein knowledge graph information during the creation of amino acid level representations. This new method integrates information at the level of individual amino acids and the entire protein. This helps capture important relationships and dependencies between proteins and their functional annotations, giving us contextually enriched protein representations.
Introduction
Imagine being a tourist in a new city who wants to see as much of the city as possible in one week. You’d use a map to locate areas but wouldn’t understand its culture by only looking at its streets without thinking about the interlinks between neighborhoods, landmarks, and the stories of the people who live there. You might gain some insights, but you’d miss out on the intricate web of interactions that truly define the city. Similarly, understanding proteins based solely on their amino acid sequences is like studying a city’s streets in isolation. Proteins, the workhorses of biological processes, are best understood within the rich context of their interactions and functional annotations.
Specifically, this work presents the formulation of the GOProteinGNN architecture as a way to integrate protein knowledge graphs with protein language models in order to enhance the learning of representations from protein knowledge graphs.
Limitations of Current Models
The current methods mainly focus on amino acid sequences, using models like ProtBert to generate amino acid embeddings and averaging these to form protein representations. Although methods like KeAP use vast protein knowledge repositories to improve protein representation, they often simplify the complex protein knowledge graphs into mere triplets, losing essential relational information. The researchers did not want to do that. Hence, they made a model, GOProteinGNN, that addresses these limitations by learning the entire protein knowledge graph.
Methods Used
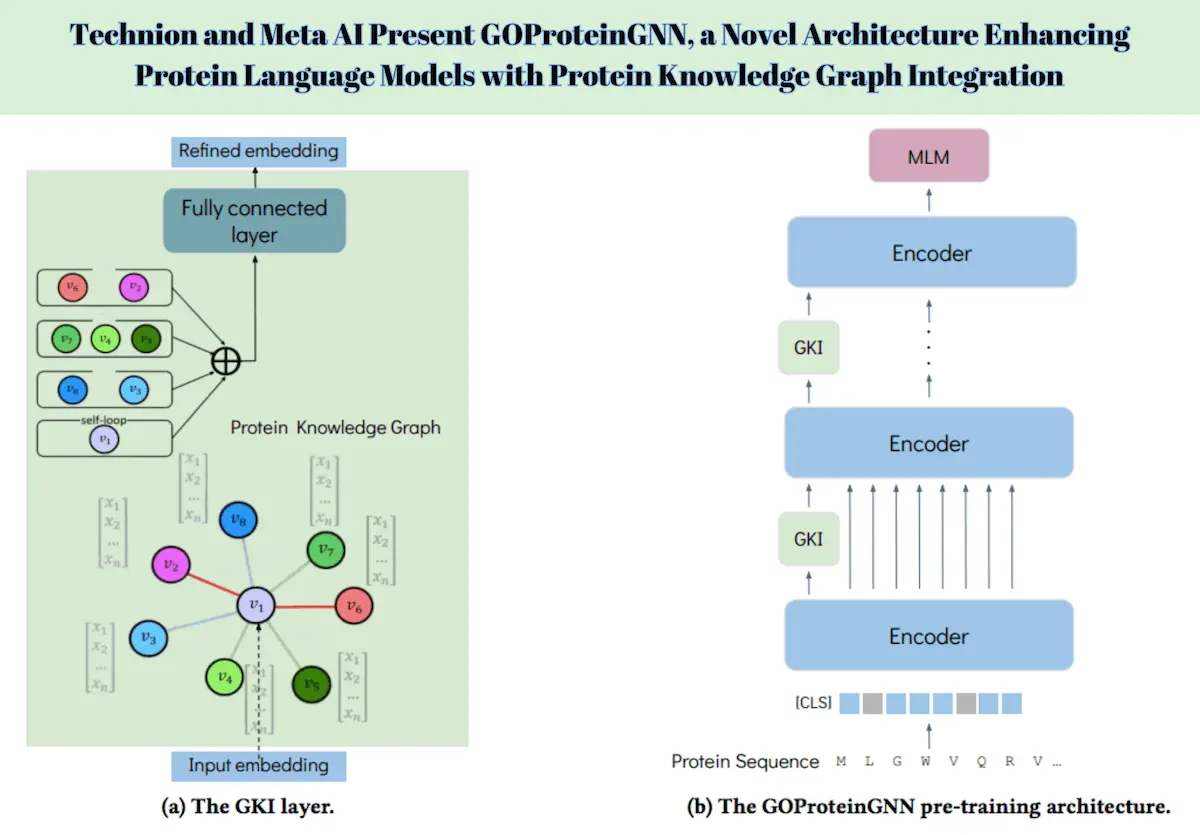
GOProteinGNN’s architecture combines both amino acid-level and protein-level representations within a single framework. It uses a novel Graph Neural Networks (GNN) Knowledge Injection (GKI) procedure that incorporates protein knowledge graph information into the [CLS] token representation of the protein language model during the encoder stage. This approach alternates between amino acid-level embedding and graph representation, which means that it gives a good encoding that integrates both sequential and graph-enhanced information.
Training Phase
Basically, the initial encoder of GOProteinGNN is a pre-trained protein language model, like ProtBert. In training, each amino acid sequence goes through several encoder layers and yields an intermediate representation. The [CLS] token representation is the complete sequence representation used to incorporate external knowledge into the framework. Specifically, the GKI layer further enriches this [CLS] token representation using the protein knowledge graph to obtain the enhanced protein embeddings that encode the vital biological contexts and interactions.
Inference Phase
During inference, GOProteinGNN introduces a weight-less injection strategy to solve the issue of dependency on a knowledge graph. This approach ensures that the enriched protein representations can be effectively used even in the absence of a knowledge graph, maintaining the model’s performance and applicability across various tasks.
Results
Empirical evaluations across numerous protein tasks show how much GOProteinGNN is better than state-of-the-art methods. The architecture outperforms existing models in various benchmarks. (On seeing some examples: GOProteinGNN gives the highest score of 0.30 in a remote homology detection task while ResNet achieves 0.17, Transformer achieves 0.21, and ProtBert achieves 0.29.) The comprehensive integration of knowledge graph information into protein language models significantly enhances the representation learning process, providing robust and contextually enriched protein embeddings.
Discussion
GOProteinGNN sets itself apart from traditional methods by uniquely encompassing both amino acid-level and protein-level representations within a single framework. This holistic approach allows for a more accurate and comprehensive understanding of protein interactions and functionalities. By leveraging the entire protein knowledge graph, GOProteinGNN captures broader relational nuances and dependencies, providing a more detailed and accurate representation of proteins.
The GNN Knowledge Injection (GKI) procedure is a key innovation in GOProteinGNN, enabling the integration of knowledge graph information into protein embeddings during the encoder stage. This novel approach enhances protein representations by incorporating sequential insights and graph-enhanced knowledge, ensuring a thorough and contextually enriched encoding.
Conclusion
Compared with previous models, GOProteinGNN indeed achieves a breakthrough in the method of learning protein representation and provides a new idea for incorporating protein knowledge graph data into protein language models. Enriched in context and anatomically detailed, this architecture effectively captures dependencies many methods fail to uncover. Therefore, through significantly outcompeting the state-of-the-art methods in protein representation learning tasks, GOProteinGNN will soon be recognized as a solution of choice with practical applications in related domains like drug discovery and bioinformatics.
Possible Future Directions
Further studies may look at the generalization of GOProteinGNN for other bodies and interactions involved in the molecular biology process and expand the outlined framework for a wider scope of biological knowledge. Also, the improvement of the GKI procedure and interaction of the presented model with other effective protein language models can improve the presented model and expand its application field. The possibility of incorporating different sources of biological information, including transcriptomics and metabolomics data, gives the prospect of further research and the evolution of the identity of protein representation learning.
Article Source: Reference Paper | GOProteinGNN is available on GitHub.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Neermita Bhattacharya is a consulting Scientific Content Writing Intern at CBIRT. She is pursuing B.Tech in computer science from IIT Jodhpur. She has a niche interest in the amalgamation of biological concepts and computer science and wishes to pursue higher studies in related fields. She has quite a bunch of hobbies- swimming, dancing ballet, playing the violin, guitar, ukulele, singing, drawing and painting, reading novels, playing indie videogames and writing short stories. She is excited to delve deeper into the fields of bioinformatics, genetics and computational biology and possibly help the world through research!