In drug development, the accurate prediction of RNA structures plays a pivotal role. Leveraging the power of machine learning, particularly Variational Autoencoders (VAEs), has emerged as a game-changer in this domain. Recent advancements by Quantori LLC, Cambridge, USA researchers have showcased the superiority of VAE models in RNA folding, surpassing traditional methods with remarkable precision.
Variational Autoencoder (VAE)
A Variational Autoencoder is a neural network model category that belongs to the generative models in machine learning. It comprises the encoder net that transforms the input data into the replication of the latent space and the decoder net that restores the input data from it. VAEs are built to learn and produce new data points from the obtained distribution within the input data and are well suited to tasks such as image generation, anomaly detection, or data compression.
Applying VAEs to RNA Tertiary Structure Prediction
For RNA tertiary structure prediction, VAEs are an innovative approach since they map the sequence context to recognize interdependencies inside strings. Using the learned representation from VAEs, scientists can predict the actual, incredibly complex 3D arrangements of RNA strands with a greater degree of fidelity than before. The generative capability and understanding of data by VAEs to describe them in a latent space allow the prediction of nucleotide positions of about 3.3 Å RMSE, thereby demonstrating the usefulness of these structures for improving knowledge about RNA and contributing to therapeutic design.
Model Details
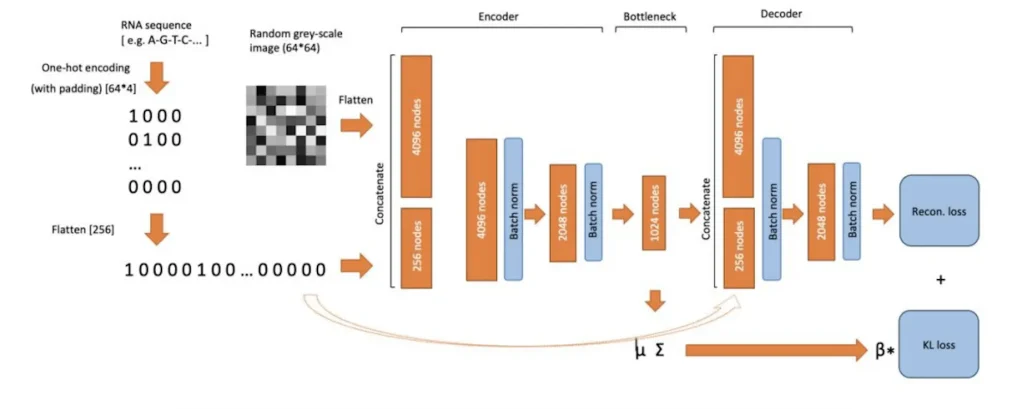
Based on the general structure of autoencoder models, VAE models for RNA structure are erected from these layers: encoder, bottleneck, and decoder. These models use a loss function that has both RMSE and KL divergence, enabling them to achieve high performance in predicting the RNA structure complexities.
Benefits of Using Variational Autoencoders (VAEs)
- Complex Relationship Modeling: VAEs are especially suited for tasks where the relationship between the data points is intricate and, in many ways, non-linear, for example, RNA tertiary structure prediction due to their ability to model higher-order structures and structures of data.
- Generative Modeling: The convolutional VAEs provide the advantage of simultaneously learning the input data distribution, thus allowing researchers to test and generate new RNA structures for analysis competently.
- Latent Space Representation: VAEs extract a low-dimensional meaningful vector, which is informative, enabling efficient data compression and traversal through different data samples by manipulating the latent space vector.
- Enhanced Prediction Accuracy: Through VAEs, researchers can receive higher predictions and thus have potentially higher accuracy in RNA structure prediction compared to typical models or algorithms, contributing to broader opportunities for enhanced biological discoveries and drug development in the future.
Real-World Applications of Variational Autoencoders (VAEs)
- Drug Discovery: VAEs are employed in the drug discovery process, which aims to obtain updated edifices of a molecule to possess the characteristic attributes that are the target of discovery.
- Image Generation: VAEs are used in various fields of computer vision work, such as image generation, reconstruction, and style transfer, and they help synthesize images of high quality with the help of learned representations.
- Healthcare: VAEs contribute to various healthcare applications, including but not limited to medical imaging and diagnostic systems, and create personalized treatments by capturing the most relevant features of the input data to impact the patient’s health.
Challenges and Future Directions
Data Scarcity: The scarcity of well-converged RNA structural data is a bottleneck for achieving well-trained VAE models to predict RNA tertiary structures.
Model Complexity: The complex shapes of RNA structures and the demands in terms of structural representation and choice of model rendering that cannot be solved using alignment information stress crucial improvements in VAE.
Incorporating Biophysical Properties: Future work involves incorporating more of the features of RNA, in light of the biophysical characteristics, into the VAE models to enhance prediction accuracy and RNA biology.
Exploring New Techniques: Continuing the development research on the paid aspect, as well as experimenting with new approaches, such as ‘diffusion models’ or ‘graph neural networks,’ could also extend the advantages of VAEs for precise and efficient predictions of RNA tertiary structures.
Conclusion
The concept of Variational Autoencoders is emerging as a powerful tool and is finding application in RNA tertiary structure prediction, which was a challenging domain earlier. Hence, an extension of VAE learning can provide significant information about RNA functions to researchers and lead to progress in areas such as drug discovery, synthetic biology, and disease studies. More complex models and applications will be developed to advance this emerging field and offer expanding opportunities in RNA biology.
Article Source: Reference Paper | Algorithm to predict the RNA tertiary structure is available on GitHub.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.