A part of systems biology is ‘designing RNA structures,’ which entails planning and organizing molecules of RNA into custom 3D formations with particular uses. RNA plays a dual role in cells, encoding genetic information and performing structural functions. Proteins are somewhat easier to study and model because of the precision nature of this macromolecule, while RNA conformations pose a special challenge because of their flexibility and multiple interactions. Deep learning and generative models have added a new generation to RNA structure design that bids new horizons for biotechnology and medicine. Enter RNA-FRAMEFLOW, a novel way for de novo 3D RNA backbone design that leverages flow-matching principles to create accurate and functional RNA structures.
The Challenge of RNA Backbone Design
The problem of designing the backbone of RNA is rooted in the fact that RNA undergoes conformational changes and because RNA interactions are complex, including base pairing and stacking. Proteins can be exemplified as relatively simple molecules by comparing them to RNA structures in which each nucleotide contains 13 atoms that require precise parameterization. This complexity, along with the loss of explicit physical interaction representations in current deep learning models, makes RNA structure prediction not only less precise than protein structure prediction but also nuanced. Solving these issues is essential for evolving the RNA modeling and design.
What is RNA-FRAMEFLOW?
RNA-FRAMEFLOW is the first generative model developed for building the backbone of 3-dimensional RNA structures. It presents a new approach by extending flow matching from SE(3), techniques usually employed in generating protein backbones, and then using them to model RNA, which is more challenging. Differing from traditional RNA structure prediction tools, RNA-FRAMEFLOW defines RNA structures as rigid-body frames and employs specific loss functions to generate realistic and diverse RNA backbones in a novel and efficient manner.
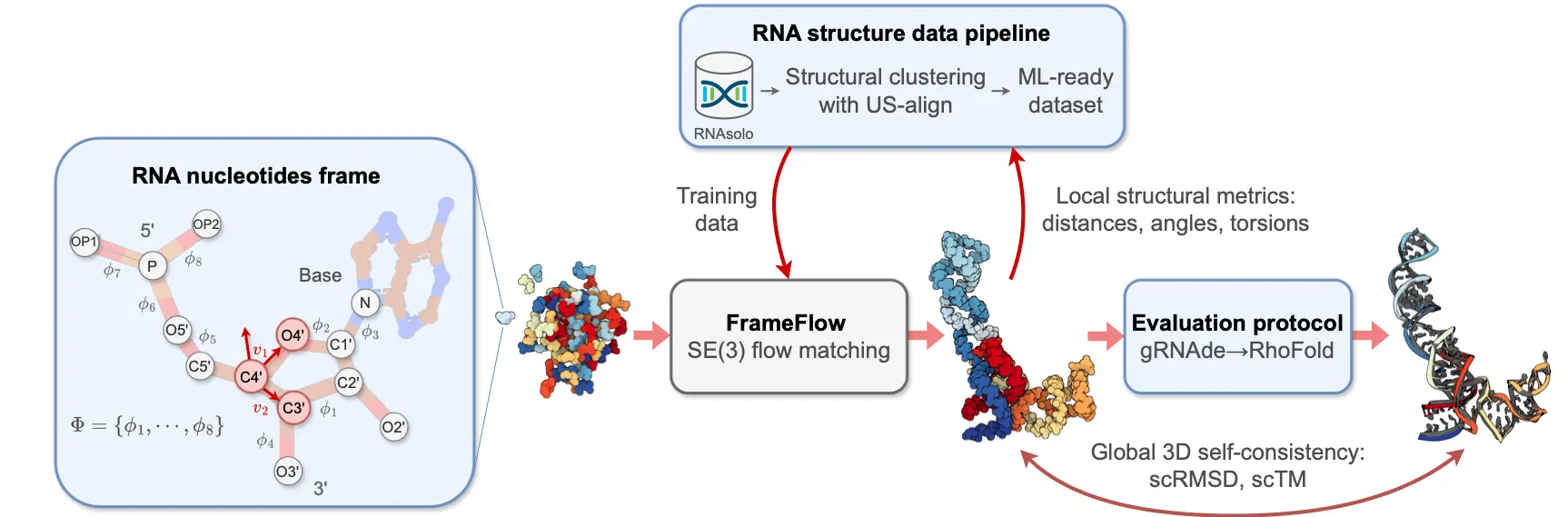
The RNA-FRAMEFLOW Pipeline
The RNA-FRAMEFLOW pipeline involves converting each RNA nucleotide into a frame to parameterize the placement of key atoms in the backbone. This process includes translation, rotation, and torsion angle adjustments to represent the 3D atomic coordinates accurately. By formulating RNA structures as rigid-body frames and implementing specific loss functions, RNA-FRAMEFLOW revolutionizes the generation of realistic and diverse RNA backbones, setting a new standard in computational biology.
How RNA-FRAMEFLOW Works?
RNA-FRAMEFLOW functions this way: it models RNA structures as rigid body frames, every single frame consisting of some atoms in the nucleotide. Based on these alterations, the targeted model calculates the location and rotation of these frames using the SE(3) flow matching of the detected features and further generates accurate 3D atomic coordinates of all the backbone atoms per nucleotide. RNA-FRAMEFLOW, which utilizes advanced loss functions and evaluation criteria for RNA modeling, further flourishes in designing locally rational and new RNA backbones, solidifying its status as a noteworthy platform in computational biology.
Advantages of RNA-FRAMEFLOW
Innovative Approach: RNA-FRAMEFLOW also proposes a new generative model restricted to RNA backbone geometry that sets the basis for the approach of flow matching with SE(3) in RNA structures.
Precision and Efficiency: Through the framework of RNA structures as rigid-body frames and with their distinct loss functions, RNA-FRAMEFLOW offers the highest accuracy and performance for creating highly relatable and diverse real-world RNA backbones.
Revolutionizing RNA Modeling: This model begins to answer questions that could have been asked five years ago, making it invaluable in computational biology and shifting the gold standard for RNA structure prediction and design.
Applications of RNA-FRAMEFLOW
Structural Biology: Using RNA-FRAMEFLOW, researchers can come up with RNA backbones with the desired 3D architecture from scratch in structural biology, which will help understand the structure-function relationship in RNA.
Drug Discovery: The capability of generating varied and authentic RNA structures by the model points directions toward drug creation and application, especially in RNA-targeted medications.
Biomedical Research: It will be quite useful in biomedical research since one can use RNA-FRAMEFLOW to examine the different structures that RNA can possess and the roles that these structures play in numerous processes in living organisms.
Evaluating the Impact
This is juxtaposed with the notion that the analytics of RNA-FRAMEFLOW does not reside in any theoretical domain since its validity and novelty are ascertained via self-consistency TM-score and novelty measures. Compared to models previously proposed in the literature, RNA-FRAMEFLOW puts itself ahead of competitors regarding the structural match of presented macromolecule backbones and takes the leading position in the field of 3D RNA backbone representation.
Conclusion
Situated in the highly complex field of Computational Biology, RNA-FRAMEFLOW is a concept that marks a turning point in the progression toward exciting new redesigns of RNA. This unified and potent generative model opens new frontiers for RNA structure designs, creating the hope for actual RNA structure probing.
Article Source: Reference Paper | Open-source code is available at GitHub
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.