The use of artificial intelligence (AI) in all phases of medication development is growing rapidly. Drug pharmacokinetic (PK) datasets are frequently acquired independently of one another, frequently with little overlap, causing data overlap sparsity, which presents a barrier for drug discovery AI. University of Waterloo researchers trying to find answers to questions in poly-pharmacy, medication combination research, and high-throughput screening find that data sparsity makes data curation challenging. Imagand is a unique SMILES-to-Pharmacokinetic (S2PK) diffusion model that may produce a variety of PK target features conditioned on SMILES inputs, according to research findings. Researchers have demonstrated that synthetic PK data generated with Imagand closely mimics the univariate and bivariate distributions of real data and enhances performance for downstream tasks. Imagand is a promising method that addresses data overlap sparsity and makes it possible for researchers to generate ligand PK data for drug discovery research in an efficient manner.
Introduction
AI is redefining drug discovery through high-throughput screening, cutting development costs, and changing ligand design and testing. Repurposing drugs, predicting drug responses, interacting with targets, poly-pharmacy, and creating artificial ligands have all been accomplished successfully using AI. The cost and duration of data gathering using assay panels are the drawbacks. Interlinking and merging pre-clinical, clinical, and chemical datasets has become simpler with advances in the standardization and distribution of these datasets. When looking for information on drug combinations and poly-pharmacy, this poses difficulties for researchers utilizing multiple datasets. Drug development must continue to advance with open data for testing and training.
Making of Imagand
Denoising Diffusion Probabilistic Models (DDPMs), which produce a new class of diffusion models capable of synthesizing ligand structures, have been used in recent developments in drug discovery AI. Diffusion models have the potential to produce pharmacokinetic (PK) features in conjunction with the ligand diffusion pipeline, as demonstrated by Hu et al. It has been shown that multi-modal diffusion models, including the Text-to-Image and Text-to-Video diffusion models, can produce excellent, photorealistic results. Additionally, Azizi et al. have demonstrated enhanced performance in ImageNet classification tasks using synthetic data from diffusion models. Motivated by these developments, scientists present Imagand, which, conditioned on learnt SMILES embeddings, can produce a variety of 12 PK target attributes from 10 PK datasets.
Key Features of the Study
- Researchers present Imagand, a cutting-edge multi-modal SMILES-to-Pharmacokinetic (S2PK) diffusion model that can produce a variety of target attributes based on SMILES embeddings that have been learnt.
- In order to facilitate training, researchers have developed a noise model that generates a prior distribution that is closer to the genuine data distribution.
- Researchers demonstrate that the Imagand model produces synthetic data that enhances performance on downstream tasks and closely reflects the univariate and bivariate distributions of the actual data.
About Imagand
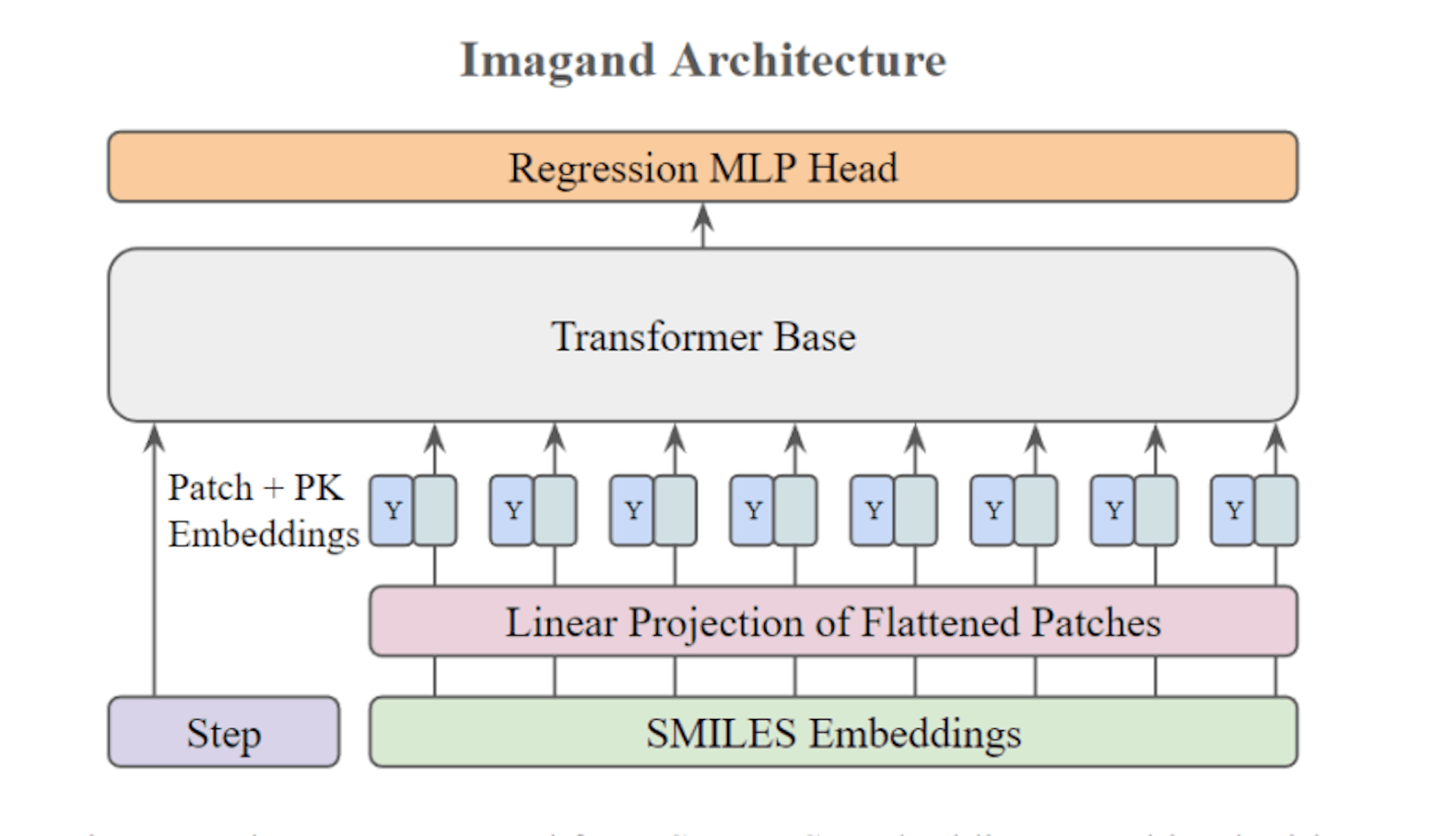
Imagand’s architecture is similar to a standard vision transformer. Using SMILES embeddings that are learnt from SMILES encoder models as a basis, Imagand is an S2PK diffusion model that generates target potential knowledge. The drawbacks of sparse PK datasets with little overlap are solved by Imagand, which produces dense synthetic data. At a fraction of the expense of performing in vitro or in vivo PK assay panels, researchers can use Imagand to construct massive synthetic PK assays spanning thousands of ligands to address poly-pharmacy and drug combination research problems.
What is Denoising Diffusion Probabilistic Models (DDPM) ?
Diffusion techniques, such as Denoising Diffusion Probabilistic Models (DDPM), are employed to model intricate datasets for learning, sampling, inference, and assessment that can be accomplished digitally. These models have been applied to medication development; they comprise a forward process of methodically destroying the structure in the data and a reverse process of learning to restore it from noise. Graph networks can be integrated with DDPMs, and small-molecule graphs with distributions resembling those of actual small molecules can be produced by Conditional Diffusion models, which are based on discontinuous Graph Structures (CDGS). Graph-level attributes and spectral and molecular features are leveraged by Digress, a graph transformer and discrete diffusion hybrid that enhances training and sampling performance.
Hindrance caused by PK properties
Understanding the body’s reaction to a drug, including absorption, bioavailability, distribution, metabolism, and excretion, requires a solid understanding of pharmaceutical kinetics (PK). PK computational methods with high accuracy are essential for small-molecule drug development. Physiologically-based pharmacokinetics (PBPK) provides PK property modeling through human body-modeled mathematical equations; nevertheless, it is unsuitable for high-throughput screening across large numbers of ligands and is dependent on costly in-vitro and in-vivo investigations. It can be expensive to extend many PK properties across large arrays of ligands, which results in minimal overlap between gathered datasets. This creates obstacles for researchers who wish to address questions that need information from multiple datasets, like drug combination research and poly-pharmacy.
Conclusion
Synthetic PK target property data that closely mimics accurate data is produced using the SMILES-to-Pharmacokinetic model Imagand for both downstream tasks and univariate and bivariate distributions. Imagand solves the problem of sparse overlapping PK target property data, enabling researchers to produce data for high-throughput screening and address challenging research topics. Imagand will scale to more datasets and larger model sizes in future work and extend to categorical PK characteristics.
Article Source: Reference Paper | Code available on GitHub
FAQ
Pharmacokinetics is the study of drugs’ movement within the body, including the processes of absorption, distribution, metabolism, and excretion. It examines how the body affects the drug, determining the time course of drug concentration and the relationship between drug dose and the resulting concentration at the site of action. Understanding pharmacokinetics is crucial in drug development and clinical practice, as it helps optimize drug dosing regimens, predict drug-drug interactions, and ensure the safe and effective use of medications.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.