Owing to the rise of global food insecurity, several initiatives have been made to understand the disease-causing plant pathogens and crop failure. Researchers from the Banfield group at the John Innes Centre and collaborators have elucidated how rice blast fungus ZiF (Zinc finger) effectors bind a conserved Exo70 interface to manipulate plant exocytosis.
Introduction
Throughout the years, global food insecurity has risen due to an increased plant pathogen and associated diseases causing crashes in yields. More than a decade of research and effort has been put into providing a better yield of crops and feeding the population. One such initiative is by researchers from the Banfield group at the John Innes Centre in collaboration with the Iwate Biotechnology Research Centre in Japan and The Sainsbury Laboratory in Norwich to describe the structural basis of effector protein (AVR-Pii) used by the blast fungus Maganaporthe oryzae to bind the rice host receptor protein (Exo70) to manipulate the plant exocytosis.
Researchers have utilized the background of understanding exocytosis. During the process of exocytosis, a protein complex exocyst mediates the binding of vesicles. The entire complex is an octamer, conserved throughout the diversity of plant kingdoms and fungi. The Exo 70 is single gene coded and has diversified throughout the years, posing it to be functionally diverse as well. In order to determine the putative effectors and virulence mechanism utilized by rice blast fungi, several potential proteins were genome sequenced.
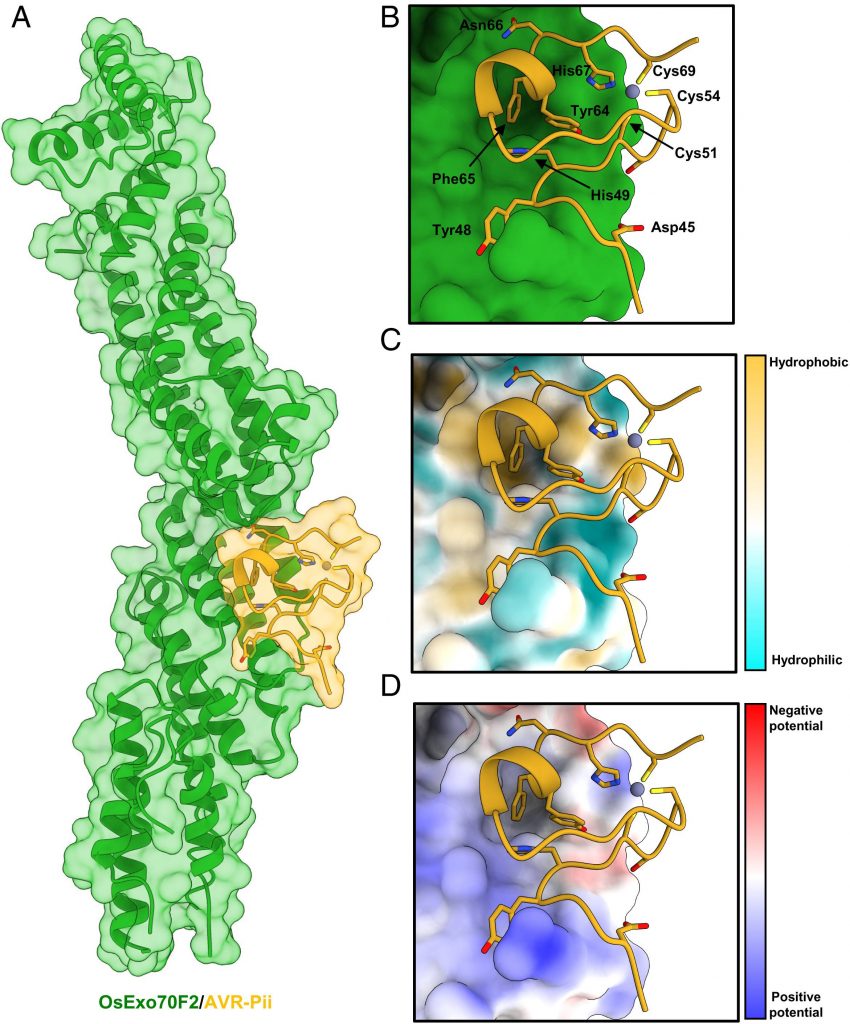
Image Source: https://doi.org/10.1073/pnas.2210559119
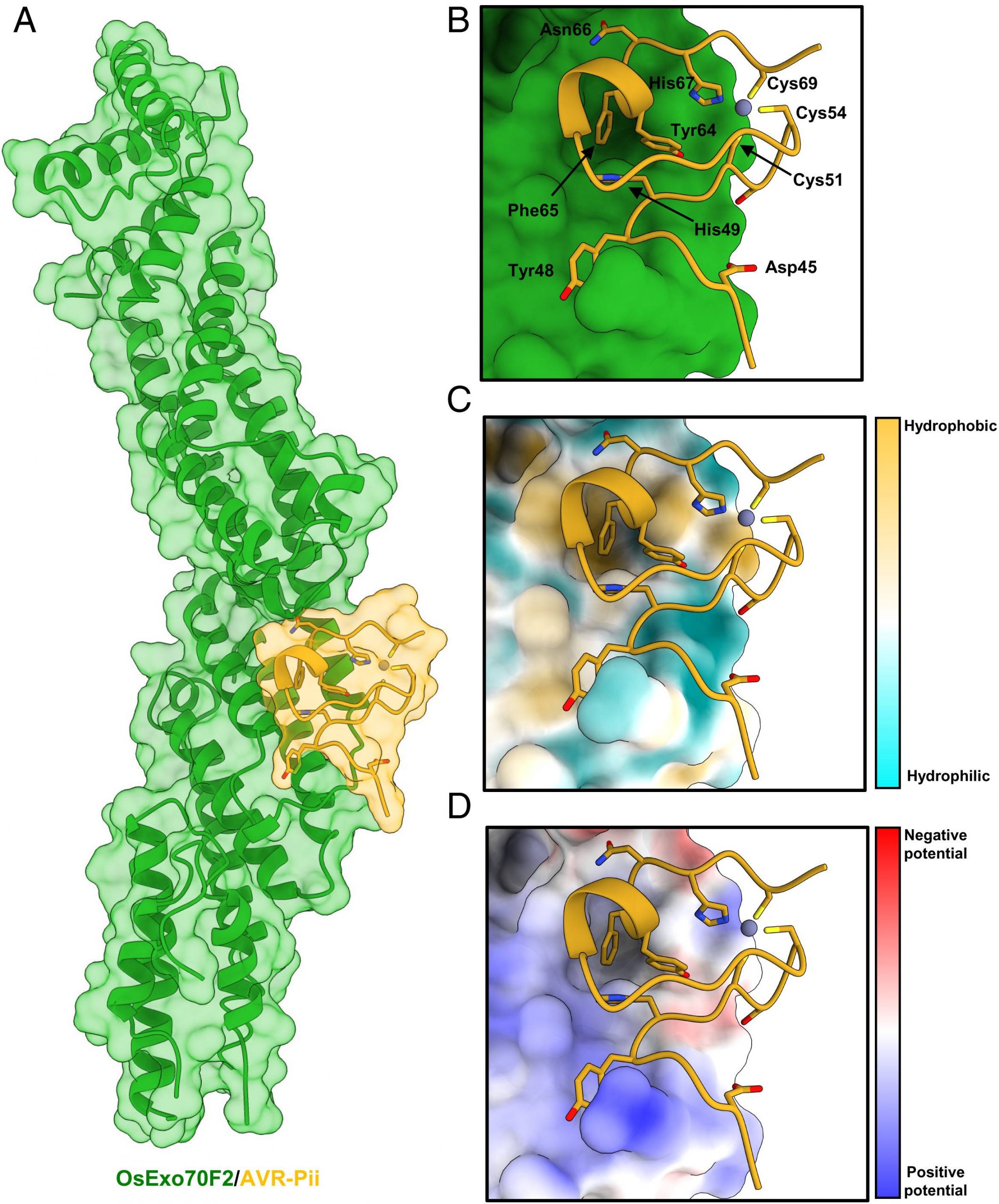
The methodology was based on illuminating the genetic basis of the M. oryzae effector protein AVR-Pii interaction with rice Exo70 proteins. The effector protein interacts with the two allele proteins of Exo 70: OsExo70F2 and OsExo70F3. The first experiment was to set to determine the crystal structure of the effector in the form of a complex with OsExo70F2. The overall results displayed the high-affinity binding of AVR-Pii with both allele proteins OsExo70F2 and OsExo70F3 performed using the Yeast-2-Hybrid assay (Y2H). The strength binding confirmed results for selective effector binding of an effector protein with the subunit proteins. The assay included yeast cells co-expressing the effector protein with the Exo 70 alleles. Followed up with validating the interactions in vitro by optimizing a pipeline to purify the Exo 70 subunits.
Secondly, researchers employed previous results to determine a crystal structure of the OsExo70/AVR-Pii complex in homogeneity. The reconstituted OsExo70/AVR-Pii complex was highly stable and was adopted to stack similarity against the fold of the Arabidopsis AtExo70A1 protein and mouse Exo 70 protein from a specific PDB database. Post placement, the team built 44 to 70 effector residues. The C-terminal region of the blast fungus AVR-Pii region revealed the fold for M. oryzae was based on the Zinc-finger motif described with supplementary data.
In order to determine the structural similarity, the search was achieved using the PDBeFold that revealed the effector protein to be similar to LIM, the structural domains of zinc fingers, wherein the previous fold with ZIF 3D structure was not reported earlier in the plant pathogens.
The team employed the understanding of 3D structure and binding to determine the nature of ligand-receptor binding in the effector protein of rice blast and OsExo70F2. Analysis
of the complex using QtPISA displayed the presence of both hydrogen and hydrophobic interaction. The most striking feature of the analysis of the OsExo70F2/AVR-Pii interface is a hydrophobic pocket formed by OsExo70F2 and residues Phe416, Leu420, Met437, and Tyr440.
The next experiment was set to determine the variation in amino acids in the OsExo70 Hydrophobic Pocket, which determines and assists the specificity of AVR-Pii binding. The analysis of the binding interface was performed using Consurf, a computational tool to estimate the evolutionary conservation of amino/nucleic acid positions in a protein or DNA molecule based on the phylogenetic relations. The results displayed unexpected conservation in the effector protein residue surrounding the hydrophobic pocket.
The modeled rice subunit OsExo70B1 using AlphaFold2 inferred the differences in binding interface residue forming the pocket with OsExo70F2 Phe416, Val419, Leu420, and Met437 replaced by Leu405, Leu408, Ile-409, and Ile426. Hence, the results concluded that AVR-Pii specificity is encouraged by small changes within the effector protein, the Exo70 binding interface, that dramatically alters the binding affinity. The following results were also applicable to the mutations at the Exo70/AVR-Pii interface that prevent the binding. The team utilized random mutagenesis accompanied by a yeast 2- hybrid assay to determine the effector protein residues involved in binding. The results supported targeted binding specificity based on the AVR-Pii residues.
Final Thoughts
Plant exocytosis has recently emerged as a preserved eukaryotic trafficking pathway with a role in both plant immunity and symbiosis. The exocyst complex formed during exocytosis is likely targeted by the effector’s proteins from diverse pathogens, and conserved Exo70 domains have been incorporated into the plant’s NLRs (Nod-like receptors) that likely act as effector baits for the detection of pathogens and virulence mediation by the immune system. Hence, the research group effector provides insight into the host-pathogen interaction, elucidating the effector protein interference with a function related to a unique role of the Exo70 subunit alleles and providing future research directions in engineering disease resistance plants.
Article Source: Reference Paper
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.