In computational biology, the ability to predict and design protein structures with atomic precision has been a long-standing goal. Proteins, as the fundamental components of life, perform many functions driven by their complex three-dimensional structures. Interactions are typically mediated through their side chains, but building an all-atom generative model requires an effective method to handle the combined continuous and discrete aspects of proteins, as represented by their structure and sequence. Alexander E. Chu and his colleagues from the University of California, San Francisco, introduced a pioneering model named Protpardelle that represents a significant leap forward in this field. This model addresses the intricacies of protein structure and sequence design, integrating sidechain interactions in a novel and comprehensive manner.
The Importance of All-Atom Protein Models
Proteins are composed of amino acids, each featuring a specific side chain that extends from the protein backbone. These side chains are crucial for the protein’s functionality, influencing everything from enzymatic activity to molecular interactions. Traditional protein design models often simplify these interactions by focusing primarily on the backbone structure, which can lead to inaccuracies in predicting protein behavior and function. The need for models that can accurately represent the sidechain interactions at the atomic level has thus been a critical area of research.
Introducing Protpardelle
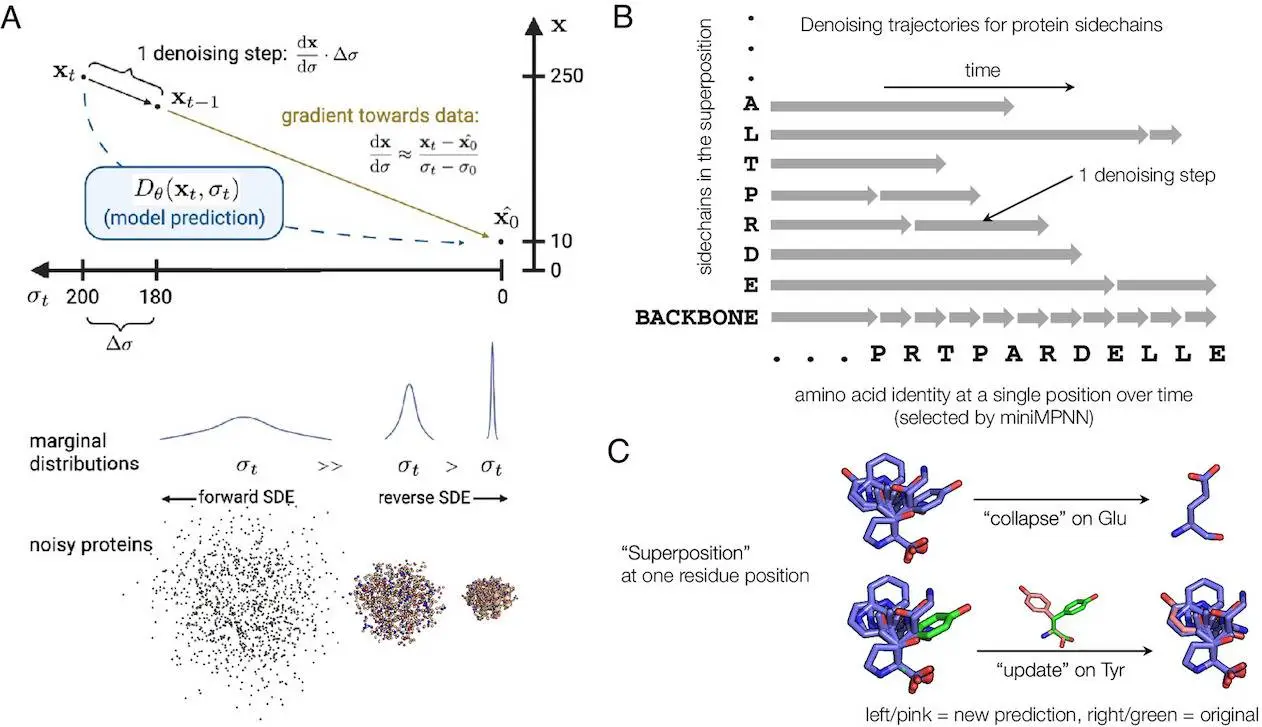
Protpardelle is an all-atom diffusion model designed to simultaneously represent (using superposition) all possible sidechain states of a protein. During the sample generation process, these superpositions collapse into specific residue types and conformations, allowing for a detailed and accurate representation of the protein’s structure. This method enables the model to co-design the backbone, sequence, and side chains of a protein, ensuring that the generated structures are both chemically and functionally coherent.
Diffusion-based Generative Modeling
Diffusion or score-based generative models have become a powerful tool for generating high-quality data samples in continuous domains, such as protein structures.
These models, including those used in protein design, employ an iterative generation process that refines samples, making them suitable for the desired application. The ability of diffusion models to incorporate new information is particularly advantageous in protein design, where the goal is to create proteins with specific properties.
In these models, forward and reverse Stochastic Differential Equations (SDEs) are defined to connect the data distribution (e.g., p0(x)) to a more manageable distribution (e.g., isotropic Gaussian distribution, pT(x)). The forward SDE reduces the signal-to-noise ratio, transforming data into whitened noise, while the reverse SDE reconstructs realistic data from noisy inputs by progressive denoising them.
In this approach, noise is systematically added to protein structures, which are then gradually refined by the model to produce accurate, realistic protein configurations. This process is akin to sculpting, where a rough shape is gradually transformed into a beautiful sculpture. This iterative refinement allows the model to handle the complex interplay between the protein backbone and side chains, ensuring that the final structures are not only accurate but also functional.
The use of a superposition state throughout the generation process allows Protpardelle to manage the protein’s dual continuous and discrete nature well. By representing all possible sidechain states and collapsing them into specific conformations at the final stage, the model can generate highly detailed and functionally relevant protein structures.
Protpardelle’s Broader Implications
This model represents a significant advancement in our understanding of protein dynamics and interactions. By providing a detailed all-atom representation, Protpardelle offers insights into the fundamental principles governing protein structure and function. These insights can drive further research and innovation, leading to discoveries in molecular biology, biochemistry, and synthetic biology.
Furthermore, Protpardelle’s approach to handling the complexity of protein structures could be applied to other computational biology and chemistry areas. The principles of diffusion-based generative modeling and superposition states can be adapted to study other complex molecular systems in drug discovery and material science domains.
Conclusion
The researchers from the University of California have made a significant milestone in the field of protein design. Protpardelle’s all-atom approach, combined with its innovative use of diffusion-based generative modeling, provides a powerful tool for designing and understanding proteins at a fundamental level. As computational biology continues to evolve, models like Protpardelle will play a crucial role in unlocking new possibilities in biotechnology and medicine, paving the way for breakthroughs that were once thought to be out of reach.
In summary, Protpardelle not only aids in designing accurate protein structures but also in designing proteins with specific, targeted functions. Its innovative approach to handling the complexity of protein structures and sequences marks a significant advancement in drug discovery and bioengineering fields. The ability to design proteins with atomic precision will undoubtedly lead to discoveries and applications, ultimately advancing our understanding of the molecular foundations of life.
Article Source: Reference Paper | Code available on GitHub.
Follow Us!
Learn More:
Neermita Bhattacharya is a consulting Scientific Content Writing Intern at CBIRT. She is pursuing B.Tech in computer science from IIT Jodhpur. She has a niche interest in the amalgamation of biological concepts and computer science and wishes to pursue higher studies in related fields. She has quite a bunch of hobbies- swimming, dancing ballet, playing the violin, guitar, ukulele, singing, drawing and painting, reading novels, playing indie videogames and writing short stories. She is excited to delve deeper into the fields of bioinformatics, genetics and computational biology and possibly help the world through research!
















[…] Protpardelle: Revolutionizing Protein Design with All-Atom Generative Model […]