Artemisinin, which is used as a cure for malaria, has remained an effective drug in managing the disease over the years. However, the recent activity arising from Plasmodium falciparum resistant to artemisinin, an important vital compound used in malaria treatment, is one of the most significant challenges threatening global health. Recent research published in Nature Microbiology by researchers from the Singapore-MIT Alliance for Research and Technology (SMART), in partnership with MIT, Columbia University Irving Medical Center, and Nanyang Technological University (NTU Singapore) has uncovered a novel mechanism behind this resistance: Regulation of tRNAs through post-transcriptional modifications is an additional layer of complexity in regulating gene expression in cells.
The Basics of tRNA and its Modifications
Transfer RNA (tRNA) complement is also an essential part of protein synthesis since it brings amino acids to the ribosome during translation using the messages received from mRNA about the codons available for use. This study overviews tRNA modifications, including their chemical characteristics, biological roles, and relationship to translation efficiency and protein synthesis. These modifications are significant for decoding accuracy, stress response, and disease processes, therefore emphasizing the role of tRNA modifications in preserving proteome homeostasis and regulating diverse cellular processes.
Artemisinin and its Mechanism of Action
Artemisinin is an effective antimalarial alkaloid from the sweet wormwood aromatic plant. It is an important ingredient of the artemisinin combination therapies (ACTs) used in malaria treatment. Artemisinin derives its efficacy through its action on the mitochondria organelle of the malaria parasite, the generation of free radicals that interfere with the structural integrity of the Plasmodium cell membrane and proteins of the mitochondria, hence leading to the death of the parasite. This specific mode of action of artemisinin makes it quite effective in combating the drug-resistant strains of malaria.
Tackling Artemisinin Resistance: A New Perspective
Resistance is due to genetic changes in the Plasmodium parasite, the most well-known of which is the Kelch13 gene. Occasionally, these mutations result in reduced efficacy of the drug at clearing the parasite from the system of the host. Since artemisinin resistance is a relatively recent phenomenon and the mechanisms have not been fully characterized, there is a critical need to identify the underlying causes of resistance and factors that contribute to drug action so as to develop implementation strategies to prevent or delay the development of such resistance.
tRNA Modification Reprogramming: A New Player in Resistance
New studies have revealed that the new element in resistant models is tRNA modification reprogramming. It was identified that, due to epigenetic modifications, altered tRNA modifications are involved in the adaption of organisms to environmental stresses and have an influence on tRNA stabilization, translation efficiency, and protein synthesis. Concerning the use of tRNAs in P. falciparum parasites regarding artemisinin resistance, tRNA modification reprogramming involved alterations in stage-specific protein expression affecting the parasite’s response to antimalarial drugs. This has clear implications for the previously unidentified strategy of epitranscriptomic changes that lie behind the evolution of the resistant parasites and could pave the way for how the fight against drug-resistant malaria could be advanced.
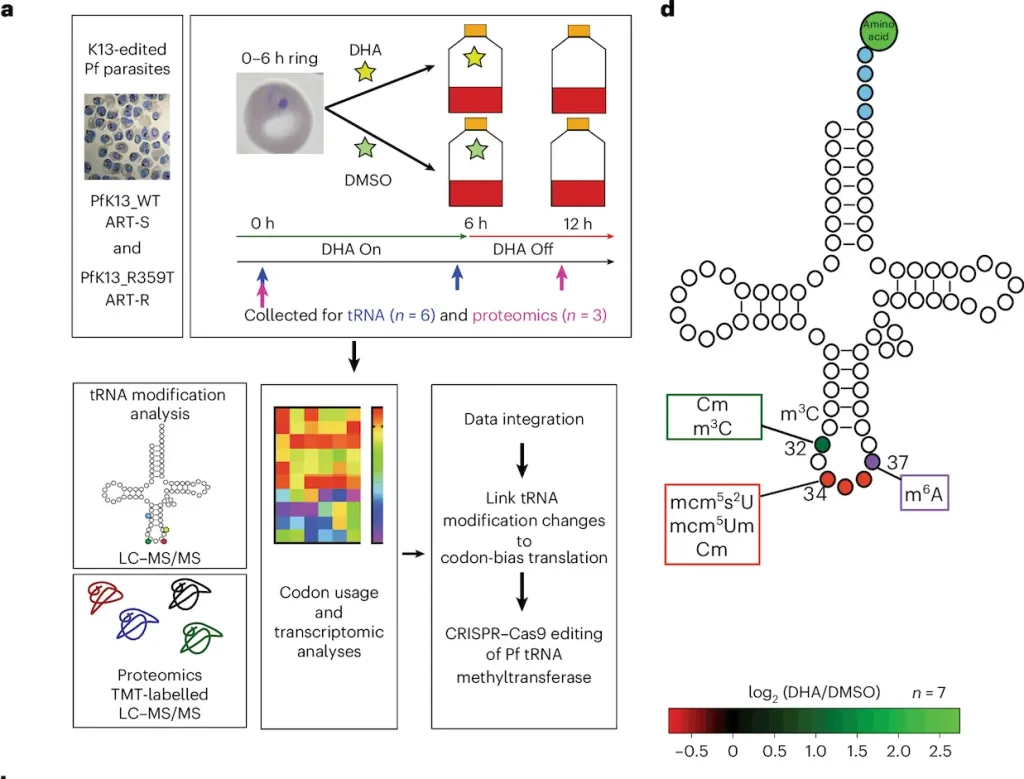
Implications for Malaria Treatment
Knowledge about the specific processes of resistance, such as tRNA translational modification profiles, has broad consequences in malaria therapy. They aim to show how parasites evolve in response to antimalarial drugs through the epitranscriptomic pathways; it facilitates the identification of specific targeting points to counter resistance mechanisms. This knowledge can inform the development of new antimalarial drugs with strategies that do not fall under the resistance of P. falciparum to increase the efficacy of treatments. Therefore, incorporating these findings as a part of effective and comprehensive clinical practice could go a long way in managing drug-resistant malaria strains and enhancing the quality of treatment outcomes in the global war against malaria.
Proteomic Analysis: Decoding the Protein Puzzle
Since the density and complexity of proteins are beyond measure in biological systems, proteomics acts as a central component in facilitating the understanding of these molecules. In the same way that identifying and weighing objects allows understanding of gravitational forces, quantifying proteins enables understanding of mechanisms occurring inside cells, interactions between cells, and disease development. Proteomic investigations can make use of sensitive methods like mass spectrometry and isobaric tagging, and they help to acquire detailed information about the proteome, effectively advancing the search for new drug targets, diagnostic markers, and therapeutic approaches. The systematic approach to learning protein attributes provides better insight into bio-complexity, especially towards disease and health management, finally leading to custom or Precision Medicine.
Methodological Marvels: LC-MS/MS Unravels the Secrets
It may be interesting to note that, out of all the analytical techniques known, liquid chromatography-tandem mass spectrometry or LC-MS/MS was indeed able to shine in decoding the most complex biological processes. LC-MS/MS derives from the marriage of the separation capability of liquid chromatography and the sensitivity and selectivity of mass spectrometry, which allows the concurrent identification and quantification of several substances: peptides and proteins. This advanced method has greatly influenced proteomic and metabolomic studies, providing profound resolutions to investigate molecular organizations, fluctuations, and social contacts in biological systems. Soon, LC-MS/MS will become essential in contemporary biomedical research since it provides high sensitivity and accuracy for new explorations in fields such as drug discovery, target identification, and individualized therapy.
Future Directions
Future research directions in malaria treatment may involve exploring innovative strategies to combat drug resistance, such as targeting tRNA modification reprogramming pathways. Additionally, advancing proteomic analyses to decipher the intricate protein networks involved in resistance mechanisms could lead to the development of novel therapeutic interventions. Integrating multidisciplinary approaches, including genomics, epitranscriptomics, and proteomics, will be essential for addressing the evolving challenges posed by drug-resistant malaria strains and improving treatment outcomes.
Article Source: Reference Paper | Reference Article
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.