The human microbiome is an essential aspect of the human body and could be responsible for several health conditions. Yet, much of microbiome transmission within and across populations and how that shapes an individual’s microbiome’s genetic makeup.is still to be understood. A recent paper in nature explores the transmission patterns of the gut and oral microbiomes between individuals via computational strain-level profiling of more than 9700 human metagenomes. The study demonstrates extensive strain sharing across individuals, with distinct transmission patterns for mother-offspring, cohabitants, and intra-populations transmission. Overall, the study’s findings underpin the relevance of bacterial transmission in human microbiome studies, specifically those on non-infectious, microbiome-associated diseases.
The Human Microbiome and the Need for Profiling Microbiome Transmission
The human microbiome comprises many microorganisms that symbiotically live on and in humans, and, more specifically, it represents the genetic makeup of these microorganisms. An individual’s microbiome is established at birth and changes over the lifetime due to various factors, including diet and lifestyle. However, since most microorganisms cannot survive outside the human body, they must be acquired from other individuals. While the gut microbiome is mainly acquired from the mother, i.e., maternal seeding, this cannot account for the vast diversity of microorganisms in adults.
However, much of how microorganisms are acquired, transmitted, and spread among populations and how this shapes the personal genetic makeup of the microbiome is unknown, especially in humans. This lack of knowledge can be attributed to the limited number and size of accurately designed studies and the challenges in accurately profiling genetic variants within strains. Although there have been some preliminary findings, there has been an utmost need for further research to understand how interpersonal relations affect the individual genetic composition of the microbiome and how it spreads within and between populations.
Furthermore, the human microbiome is composed of individual-specific strains, which can significantly vary in their genomic and functional characteristics even within the same species. Accurately profiling these strains is crucial for distinguishing between the transmission and convergence of microorganisms within an individual’s microbiome. In fact, understanding the transmission of microbiomes sheds light on the complexity of the human microbiome and helps address the communicable aspect of diseases that are currently considered non-communicable.
Recently, researchers from the University of Trento have provided a comprehensive understanding of the transmission of microbiome strains between individuals across various scenarios. The researchers characterized and quantified the patterns of person-to-person microbiome strain sharing, and by doing so, they provided further insights into the transmission landscape of the human microbiome, specifically the human gut and oral microbiome.
Profiling Microbiome Transmission
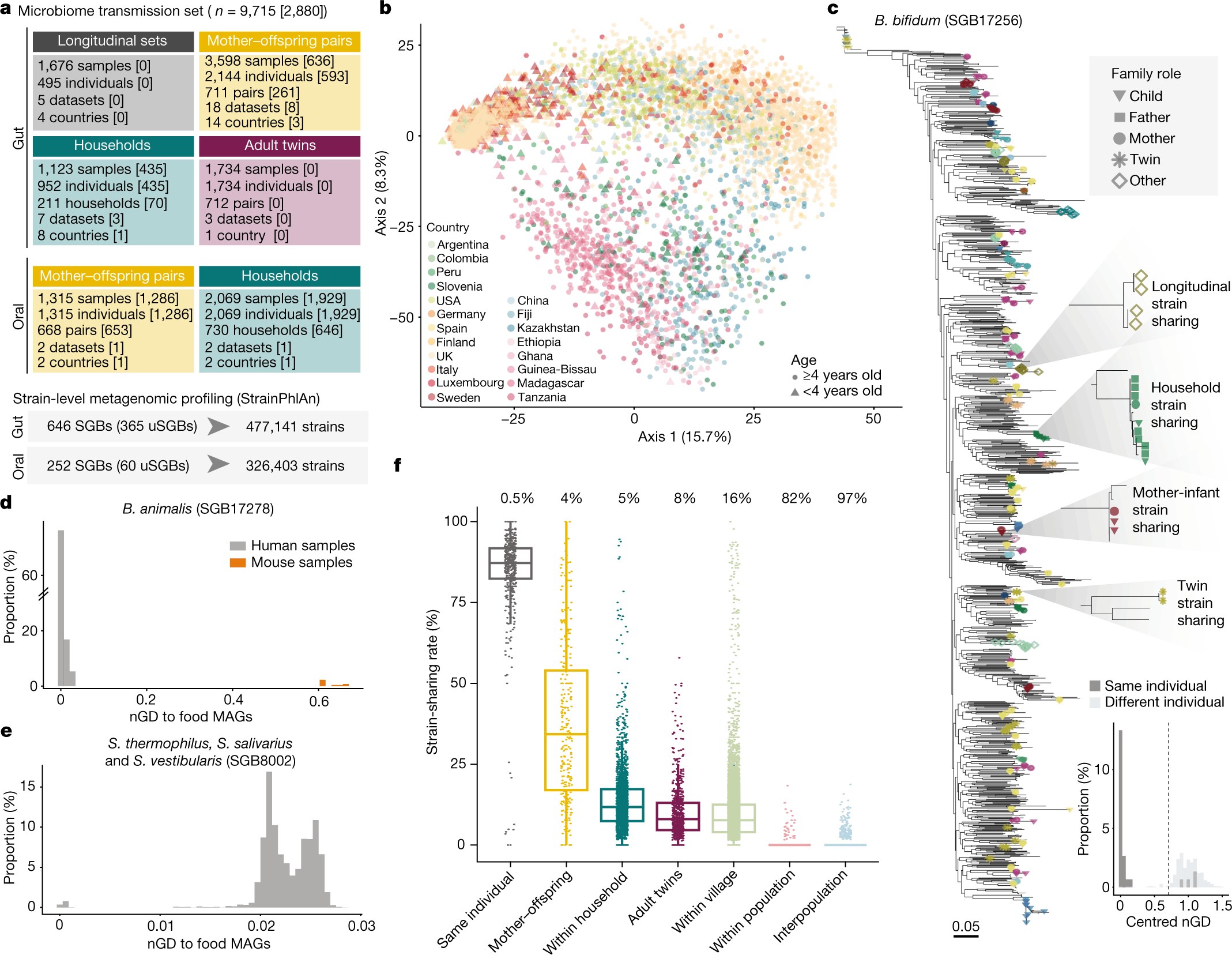
The researchers analyzed 31 metagenomic datasets with known family relationships sequenced from different geographical regions and host lifestyles to unravel the modes of person-to-person microbiome transmission. The collection comprised 9715 microbiome samples and included host information. The researchers used metagenomics to infer microorganism strain transmission and refined their strain-level profiling methodology. They also identified normalized phylogenetic distance thresholds that best differentiated same-individual longitudinal strain retention from unrelated individual nGD distributions. The findings showed that 1022 not-yet-cultured and unnamed species-level genome binds (uSGBs) were identified, constituting 37% of all detected species-level genome bins (SGBs).
Strain sharing was evaluated by profiling the dominant strain of SGBs found with a minimum of 10% prevalence in at least 20 samples in one or more cohorts. Bifidobacterium was on such gut SGB assessed for transmission, and it was seen that this strain was detected in 87% of pairs of samples from the same individual collected up to six months apart. Moreover, the study identified 646 SGBs in gut metagenomes and 252 SGBs in oral metagenomes shared between individuals. Finally, they also detected 13,278 instances of inter-individual shared Bifidobacterium bifidum strains between most mothers and their offspring.
The gut microbiome transmission profile was then analyzed to determine how microbial strains are shared between people. The researchers found that strains are highly persistent in individuals and the fact that strain-sharing rates are the strongest between cohabiting mothers and their children, followed by individuals in the same household, non-cohabiting adult twins, and non-cohabiting adults in the same geographical region (in the same village). Essentially, there was minimal strain sharing between non-cohabiting individuals in different villages or populations. These findings illustrate the relevance of that direct person-to-person interaction and social-interaction networks in dictating an individual’s gut microbiome.
Major Factors Driving Microbiome Transmission
- Mother-Offspring Transmission
The study conducted by the researchers suggests a negative correlation between the age of offspring and the rate of strain-sharing with their mother, regardless of the increase in the number of shared species over time. This is because, during the first year of life, the offspring share half of the strains of the species with their mothers found in both their microbiomes. However, as the offspring gets older, strain sharing decreases. From 1-3 years of age, strain sharing was found to be reduced to 27% but stabilized after three years of age. Finally, the individual lifestyle and the mode of infant delivery did not significantly influence microbiome transmission rates.
- Cohabitation
The study evaluated the transmission of gut microbiomes within households to see the effect of cohabitation. It was found that households with cohabiting members demonstrated substantially higher strain-sharing rates than non-cohabiting individuals, suggesting that horizontal transmission of microbiomes occurs in households. Like mother-offspring transmission, the stain-sharing rate of cohabitants decreased with age, and the number of non-family origin strains increased. Moreover, family members also demonstrated substantially higher strain-sharing rates than different-household comparisons. In addition, strain-sharing was also assessed for adult twins, and it was found that the rate of strain-sharing reduced with the number of years the twins spent living apart, again suggesting that cohabitation highly influences microbiome transmission in adults.
Conclusions
The study on microbiome transmission across diverse populations demonstrated how bacteria are passed from person to person and found that the phenomenon is more prevalent than previously thought. The study found that even though strain-sharing was most significant between mother and offspring, it could also occur between cohabitants. However, the researchers highlight the need for further research to understand highly transmissible species’ genomic and phenotypic characteristics.
Article Source: Reference Paper
Learn More:
Diyan Jain is a second-year undergraduate majoring in Biotechnology at Imperial College, London, and currently interning as a scientific content writer at CBIRT. His passion for writing and science has led him to pursue this opportunity to communicate cutting-edge research and discoveries engagingly to a broader public. Diyan is also working on a personal research project to evaluate the potential for genome sequencing studies and GWAS to identify disease likelihood and determine personalized treatments. With his fascination for bioinformatics and science communication, he is committed to delivering high-quality content a CBIRT.