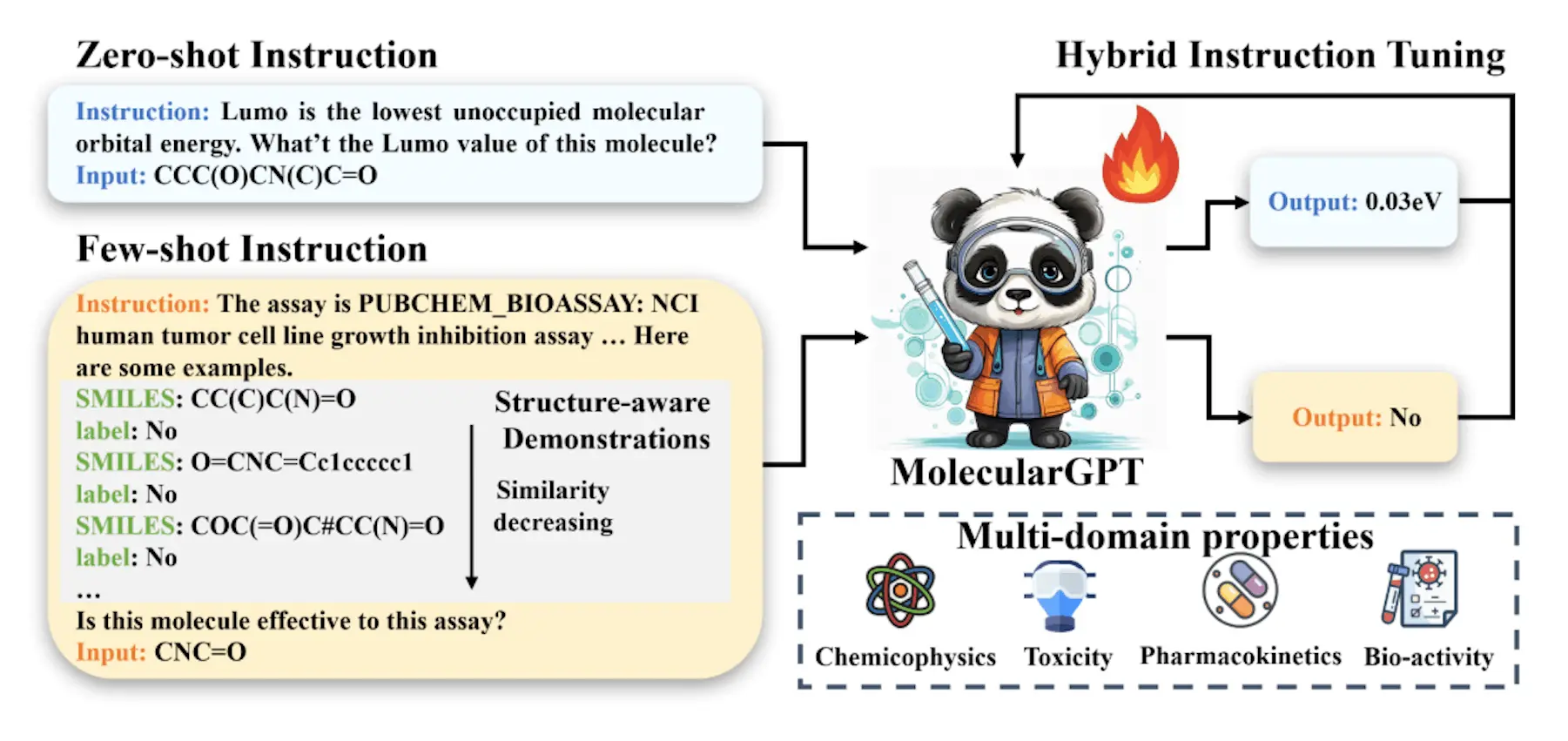
Drug discovery requires the fundamental process of molecular property prediction or MPP. However, the shortcomings of previous approaches—crucial for real-world applications—are their limited capacity to generalize for novel and unforeseen jobs and their need for a high number of labeled molecules. The researchers present MolecularGPT for a few-shot MPP to address these issues. Researchers refine large language models (LLMs) using carefully selected molecular instructions from more than 1000 property prediction challenges, emphasizing instruction tweaking. This makes it possible to create a flexible and specialized LLM using zero- and few-shot in-context learning (ICL) that can be applied to novel MPP problems without fine-tuning.
Introduction
Progress in areas like health and materials science depends on identifying molecules with desirable functional characteristics. Molecular property prediction successfully expedites the drug discovery process and lowers related expenses. MPP uses deep learning algorithms to predict the functional properties of molecules. Among them, techniques based on graph neural networks (GNNs) have produced state-of-the-art outcomes recently. These techniques, however, are only applicable in supervised environments, which runs counter to realistic requirements given the high cost and time commitment associated with molecular annotation. Additionally, the model’s generalization capacity in open-world scenarios may be limited by the task-specific supervised learning process, which hinders the model’s ability to adapt to new tasks.
Background
Studies have been done to integrate molecular representations and natural language to enable zero-shot reasoning for molecular probability models. Through contrastive learning, CLAMP, a text-molecule model, aligns chemical text and molecular graphs and classifies a query molecule’s bioactivity based on similarities. Nevertheless, CLAMP is not a generative model; rather, it is restricted to classification tasks. In another area of LLM research, task descriptions, and molecule graphs are combined into a single generative LLM to enable zero-shot reasoning for molecular property prediction in both regression and classification tasks. Nevertheless, it is constrained in its ability to produce few-shot molecular property predictions by an additional architectural design. A research question arises: Can LLMs be fine-tuned for generic MPP, allowing the model to inherit LLMs’ few-shot ICL capacity and generalize to unforeseen tasks? No LLM-based technique in the molecular domain fully inherits LLMs’ generalization and ICL abilities.
Understanding MolecularGPT
MolecularGPT is a revolutionary instructional-tuned LLM that retains its zero-shot and few-shot in-context reasoning capabilities while generalizing to various novel MPP problems. Instruction creation uses the SMILES representation of molecules as a unified graph-to-string translation, effectively converting the chemical structures of molecules into atomic symbols and chemical bonds. Structure-aware few-shot instructions are proposed, including the top-k neighbors of each molecule as complementary information for instruction creation, enabling the full utilization of graph structures in molecules. This design is readily appropriate for few-shot ICL since it follows the format of MolecularGPT’s instruction tuning and inference prompts. MolecularGPT can do well in both zero-shot and few-shot property predictions thanks to its hybrid instruction set, which combines both zero-shot and few-shot instructions.
Key Contributions of the Study
- To facilitate efficient few-shot MPP in the ICL fashion, researchers investigate how to modify pre-trained LLMs for the molecular field. The first instructionally fine-tuned LLM that facilitates few-shot property prediction on unknown tasks without the need for fine-tuning is called MolecularGPT, according to the researchers.
- To improve LLMs’ compatibility with molecular fields, researchers present the idea of structure-aware fewshot instruction. Instead of combining graph structures and SMILES representations from a model-centric standpoint as in previous efforts, researchers create global structure-aware few-shot demos to maliciously combine them from a data-centric perspective.
- Researchers create a hybrid instruction set that inherits LLMs’ capabilities for few-shot ICL. This collection yields 3.5GB of training tokens and consists of a combination of few-shot and zero-shot instructions covering over 1000 MPP problems, including both classification and regression tasks in the chemical, biological, and quantum mechanical domains. Empirical evidence has demonstrated the effectiveness of this varied instruction set in helping LLMs adapt to MPP tasks.
- The efficacy of MoelcularGPT was confirmed by comprehensive experimentation on ten molecular property standards at various scales and jobs. The empirical findings show that MoelcularGPT performs up to 16.6% better on average across all classification tasks than the leading LLM baselines, LLaMA-13B. Furthermore, MolecularGPT sets new standards for few-shot molecular property tasks by outperforming conventional supervised GNN techniques on four out of seven datasets with just two-shot samples.
Experimental Results
When two or more retrieval molecules are used, the marginal benefit of using molecularGPT models decreases, while they still exhibit notable improvement. With 512 tokens as the maximum input length and a maximum of 4 examples in the instructions, the model’s performance does not improve with additional retrieval molecules. This could be because of noise introduction. Particularly when there are more demonstrations, the descending order of similarity is preferable to the ascending order of similarity. In all classification tests, the retrieval strategy based on similarity surpasses the one based on variety, so similarly retrieved molecular demonstrations perform better than diverse demonstrations. The diversity-based technique provides a range of positive and negative examples, which may introduce noise and lead to uncertainty, whereas the similarity-based methodology more closely corresponds with query molecules.
Limitation
In this work, researchers present molecules using SMILES strings; however, they ignore geometric structural information and basic tasks such as molecule synthesis, optimization, and captioning. This constraint restricts the model’s practical applications and makes it more difficult to simulate molecules found in the actual world. For classification problems, it is compatible with supervised GNN models; however, because of difficulties in producing numbers directly for basic LLMs, regression tasks have shortcomings.
Conclusion
The model MolecularGPT has been trained on over a thousand prediction tasks and optimized for training and inference phases. It has been demonstrated to perform better on several datasets than supervised and even basic language models in few-shot scenarios. The project aims to provide LLMs, particularly LLaMA, with a broader understanding of molecular properties so that they can use zero-shot and few-shot ICL to generalize prediction problems outside their area.
Article Source: Reference Paper | MolecularGPT code is available on GitHub.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.