Genomics to Notebook (g2nb) is a new environment that combines the popular JupyterLab notebook system with well-established bioinformatics platforms to provide a seamless and accessible workflow for genomics analysis. While Jupyter Notebook and JupyterLab have become widely used in data science and bioinformatics, incorporating existing bioinformatics analysis platforms like Galaxy and GenePattern into the notebook metaphor has been challenging. g2nb aims to bridge this gap by integrating these platforms into the notebook interface, allowing users to leverage thousands of genomics methods without the need for programming.
Seamless Integration of Bioinformatics Platforms
In g2nb, a new cell type is introduced that serves as an interface to tools hosted on remote Galaxy or GenePattern servers. These analysis cells present a form-like interface similar to the original platforms, enabling users to input parameters and data for analysis. Once launched, the analysis runs on the specified remote server, and the job execution status is displayed within the cell. Upon completion, links to the result files are provided in the notebook cell, which can be easily used as input for subsequent analyses. This seamless integration within the notebook creates a cohesive analysis workflow for users.
Interactive Visualization Capabilities
The g2nb environment also includes interactive visualization capabilities for various genomics datatypes. It incorporates JavaScript versions of Cytoscape and the Integrative Genomics Viewer (IGV), as well as standard plots available in GenePattern and Galaxy. These visualizations can be displayed within notebook cells or launched in independent windows for better visibility. g2nb leverages existing functionality provided by these projects to run within JupyterLab. For example, CyJupyter and igv-notebook enable the use of Cytoscape and IGV, respectively, within the notebook environment.
Enhancements for Smooth Data Flow
To facilitate smooth data flow within the notebook, g2nb introduces several enhancements to JupyterLab. It allows users to access Galaxy histories and GenePattern result files within notebook cells, enabling easy selection of files from previous analyses. A list of all analyses in a notebook that can receive a particular result file as input is also displayed. Additionally, g2nb enables the execution of a sequence of cells as an end-to-end workflow, including cells that launch jobs on remote servers. Data transfers between remote servers and the g2nb workspace are handled automatically. The Globus file transfer protocol is integrated into g2nb, providing robust file transfers between the workspace and any Globus endpoint.
Simplified Data Transfer and Python Integration
For programmers, g2nb simplifies the transfer of data between Python objects and Galaxy/GenePattern jobs. Python variables can be provided as input parameters to analyses, and the g2nb environment evaluates the variables and passes their values to the analysis. Moreover, users can directly load the contents of result files into Python variables or Pandas dataframes, eliminating the need for manual downloads and file reading.
User-Friendly Interface Building
g2nb offers a User Interface Builder for notebook authors, allowing them to present code cells in a more user-friendly format. Authors can create web form interfaces to cells, exposing only the necessary parameters for users to input. Data inputs, text or numeric entries, and dropdown lists can be specified, tailoring the interface to the specific code. The underlying code is always accessible via a toggle button.
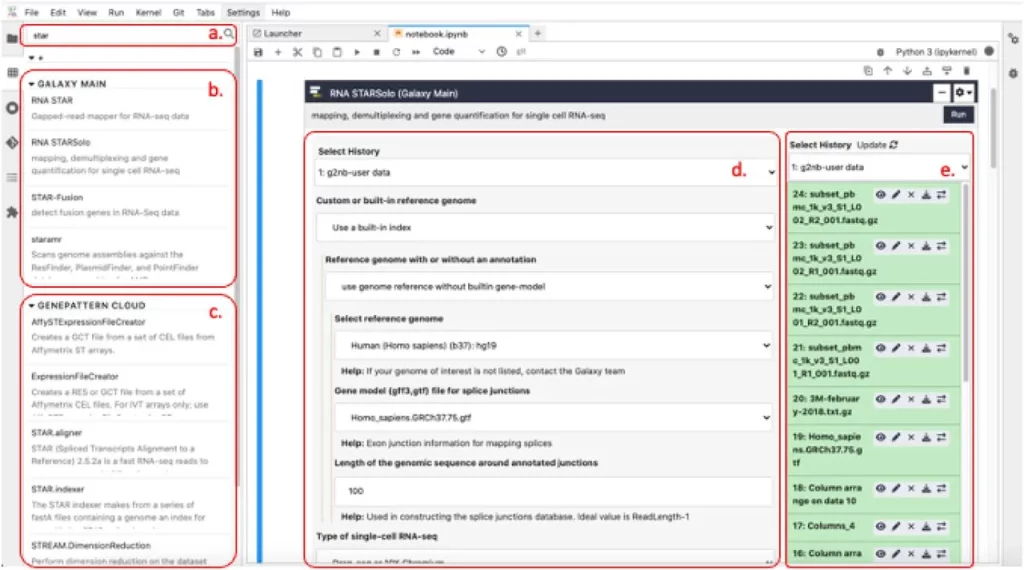
Image Source: https://doi.org/10.1101/2023.04.04.535621
Collaboration and Sharing through g2nb.org
A freely available online workspace called g2nb.org is provided to facilitate collaboration and sharing. It allows investigators to create, run, and share notebooks, as well as publish them for general use. The workspace includes a growing library of g2nb notebooks that implement common analysis workflows, serving as templates for researchers. To address issues with incompatible dependencies, g2nb offers project spaces, which provide separate contexts containing notebooks, packages, libraries, and files. Projects can be shared with collaborators and published on the g2nb workspace.
Conclusion
Genomics to Notebook (g2nb) is a unique environment that combines the JupyterLab notebook system with popular bioinformatics platforms, such as Galaxy, GenePattern, and the JavaScript versions of IGV and Cytoscape. By integrating these platforms into the notebook interface, g2nb offers thousands of genomics methods without the need for programming. Overall, g2nb stands out as a comprehensive notebook-based system that incorporates multiple bioinformatics analysis platforms, making it a valuable tool for researchers across various domains.
Article Source: Reference Paper | g2nb: Platform | Source Code
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.









