In a groundbreaking study from the Whitehead Institute for Biomedical Research and MIT’s Computer Science and Artificial Intelligence Laboratory, researchers have uncovered a hidden layer of biological organization—protein sequences contain codes that determine their precise location within a cell. This study introduces ProtGPS, an AI-driven model that predicts and manipulates protein localization with unprecedented accuracy.
How Do Proteins Find Their Location?
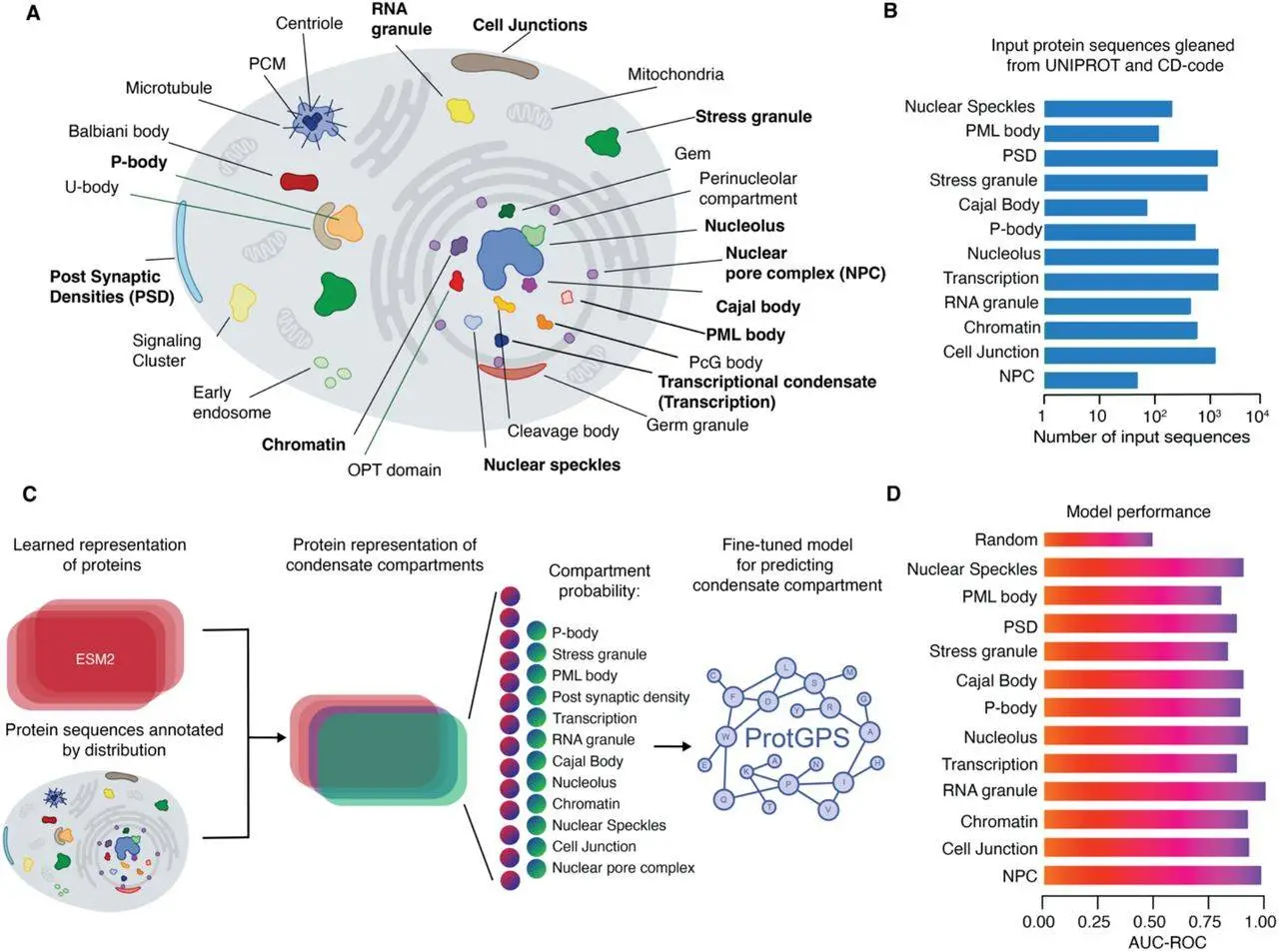
Proteins operate in specific predetermined regions inside a cell. Before, scientists assumed these compartments were reached mainly through structures. This investigation indicates that protein sequences, by themselves, also contain instructions for reaching the correct compartment, such as for the nucleus, nucleolus, or nuclear speckles. Knowing the mechanisms for sorting proteins in a cell is critical in revealing new biological systems and in formulating new therapeutic approaches.
The ability of proteins to localize correctly is very important for a cell’s proper functioning. Localization problems may cause severe diseases such as neurodegenerative disorders and various cancers. The study of these phenomena is important for understanding the internal processes existing in a living cell and the pathological disorders occurring in it.
ProtGPS: A Tool for Protein Localization
ProtGPS is a deep learning model that has been trained on more than 5,500 human proteins. It takes amino acid sequences as input and makes predictions about the subcellular localization of a protein by identifying several patterns that determine its suitability to various compartments. The model reaches accuracy scores from 83% to 95% across the partitions of a cell in 12 compartments.
The employment of artificial intelligence-enabled tools, like ProtGPS, allows better classification of proteins than traditional experimental methods, which are costly and slow. This approach allows researchers to investigate protein interactions in much greater detail and on a bigger scale. Moreover, ProtGPS can also help analyze the impact of mutations on the subcellular localization of proteins, which can help understand some genetic disorders.
Designing New Proteins
Employing a computational technique known as Markov Chain Monte Carlo (MCMC), researchers created synthetic proteins that were capable of selectively self-assembling within the predetermined compartments. When tested on human cells, the proteins, for the most part, efficiently localized to the correct places; thus, ProtGPS was successfully able to demonstrate that it works as intended.
This ability to design synthetic proteins that can be targeted to specific locations is a significant achievement in molecular biology. It is now possible to construct proteins that efficaciously perform in particular cell settings, which can be beneficial for synthetic biology, biotechnology, and even therapeutic development. For instance, proteins that target cancer cells can be designed to release their cytotoxic drugs precisely where they are needed. This will greatly reduce the side effects of such drugs and increase the effectiveness of the treatment.
Conclusion
This study shows that proteins have inherent localization codes apart from their structural ones. ProtGPS and similar tools give useful information about cellular organization and hold promise for growth in biotechnology and medicine. Understanding protein localization using AI is creating new opportunities for treating diseases and advancing synthetic biology.
With the acceleration of AI-powered research, the discoveries in protein biology and cell organization are likely to increase. More sophisticated protein prediction and design models, enhanced abilities to synthesize proteins for unwanted cellular mislocalization, and cure-easing targeted mislocation of proteins are some of the possibilities. Unlocking the secrets of targeting and changing the position of proteins will greatly influence genetics, medicine, and biotechnology in the next few years.
Article Source: Reference Paper Abstract | Full Paper on bioRxiv | Reference Article.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.