Myeloid neoplasms are linked to mutations or deletions of the U1 snRNP-associated factor LUC7L2, and cellular metabolism is changed when LUC7L2 is knocked down. Here, researchers from MIT demonstrate that members of the LUC7 protein family widely influence mRNA splicing in leukemias with LUC7L2 copy number variation as well as human cell lines, and they differentially regulate two key classes of 5′ splice sites (5′SS). Based on the study, splicing of “right-handed” 5′SS in humans is stronger on the intron side of the near invariant /GU, whereas splicing of “left-handed” 5′SS is enhanced by LUC7L2 and LUC7L with stronger consensus matching. Human LUC7 protein splice site mutations and domain swapping were used to validate this notion of sequence-specific 5′SS regulation. According to an evolutionary study, the LUC7L2/LUC7L3 subfamilies existed prior to the division of plants and animals.
Introduction
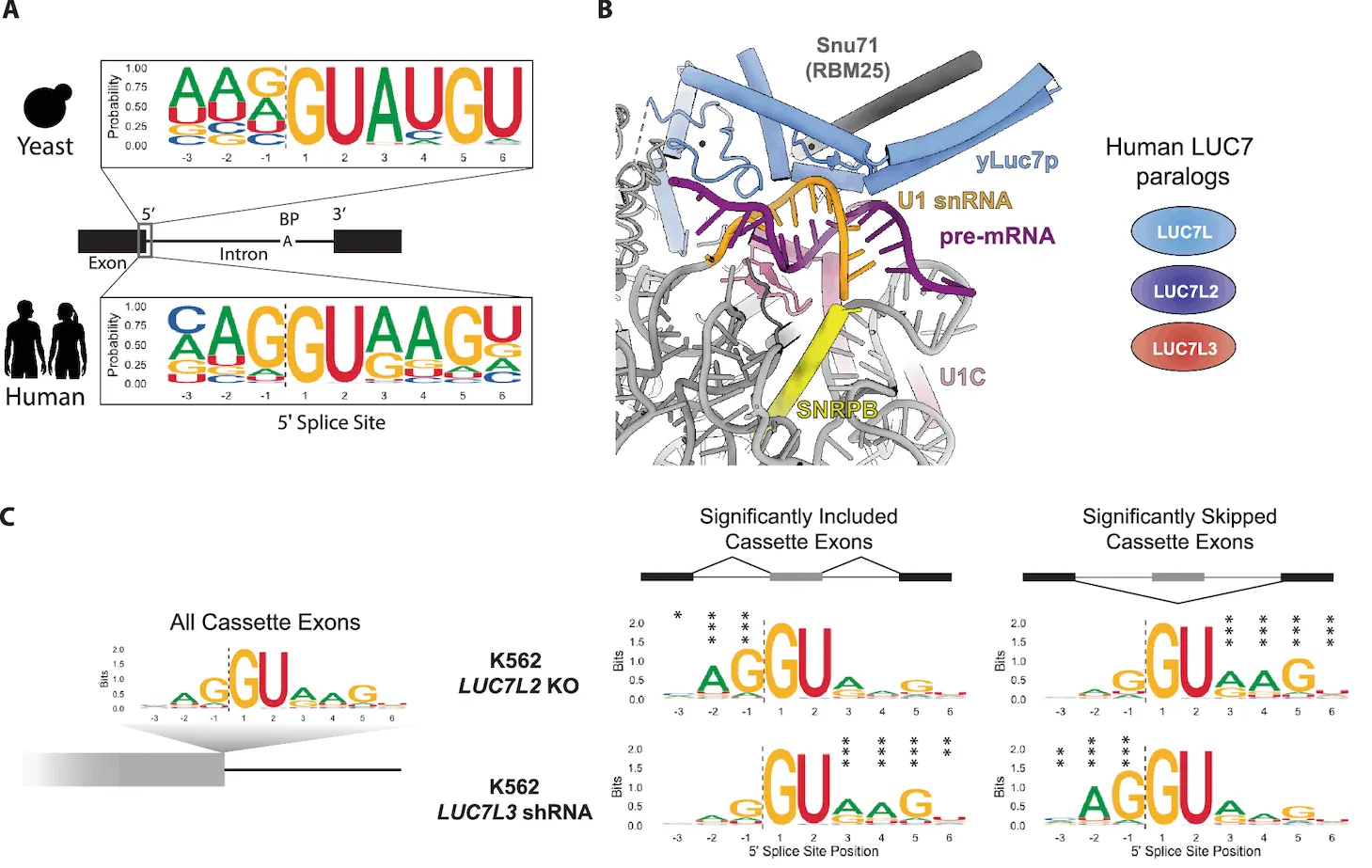
In eukaryotic protein-coding genes, splicing is accomplished by spliceosome assembly, in which the U1 snRNP uses RNA: RNA base pairing between the 5′ end of U1 snRNA and the 5′ splice site (5′SS) to detect putative intron boundaries in pre-mRNAs. Significant differences in functional 5′SS motifs between eukaryotes indicate that other factors play a role in the recognition of pre-mRNA splicing substrates despite the 5′ end being almost invariant.
In erythroleukemia cells, a surprising link between mRNA splicing and cellular metabolism was found. Growth in galactose-containing media was aided by the enhanced use of oxidative phosphorylation (OXPHOS) brought about by genetic ablation of U1 snRNP components. Changes in important metabolic genes like PFKM and SLC7A11 helped partially explain how the deletion of LUC7L2 changed the splicing of several hundred exons.
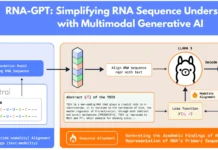
During the early phases of pre-spliceosome assembly, yeast LUC7 interacts with the phosphodiester backbone of U1 snRNA, according to cryo-EM investigations of the S. cerevisiae spliceosome. Mammals, in contrast to yeast, have three LUC7 proteins: LUC7L, LUC7L2, and LUC7L3. U1 snRNP proteins were obtained by immunoprecipitation of each human LUC7 protein, demonstrating their association with the snRNP. Splice sites in several pre-mRNAs and nucleotides at the 5′ end of U1 snRNA that base pair with the 5′SS cause all three proteins to crosslink. Human LUC7 proteins are not present in any of the known cryo-EM spliceosome structures despite their conservation and influence on the transcriptome.
The Effect of LUC7 Paralog-Specific Splicing Regulation on Leukemogenesis and Hematopoietic Differentiation
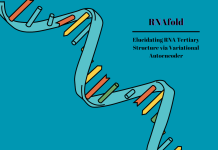
The functional differences among LUC7 paralogs affect splicing and differentiation in hematopoietic stem cells produced from induced pluripotent stem cells. LUC7L2 deletion or low expression via del(7q) affects differentiation and is linked to the emergence of leukemia and other myeloid neoplasms. Nevertheless, nothing is known about the molecular roles of these paralogs. Several human LUC7 paralogs have a predictable, 5′SS-sequence-dependent effect on the splicing of thousands of exons, according to a study examining the unique roles of LUC7s in pre-mRNA splicing. The different phenotypes of “left-handed” (LH) and “right-handed” (RH) 5′SS are explained by the opposing ways in which two subfamilies of LUC7 proteins regulate these two classes of paralogs.
Damage to the LUC7L3 Subfamily is Linked to Changes in the Composition of 5′SS.
As stated in the findings, the LUC7L3-subfamily genes in humans and A. thaliana promote LH 5′SS and RH 5′SS, indicating that these subfamilies developed before mammals and fungi split from plants. Given that none of the fungal species under study had an ortholog for LUC7L3, it is likely that this subfamily vanished from the fungal lineage shortly after it split from the animal line. The majority of fungal genomes today have developed primarily without the LUC7L3 protein. Distinct patterns of information content between the 5′SS of plants, animals, and fungi were found in the investigation of 33 eukaryotes, which represent a variety of plants, animals, and fungi. While there is little to no bias at any exonic location, the makeup of all examined fungus is heavily biased on the intron side of the 5′SS.
In both exonic and intronic places, animals exhibit moderate biases, with the exception of three protostomes (insects and worms). Although plant 5′SS are more like animals than fungi, they do have a slightly higher bias on the exon side than mammals. With the loss of the LUC7L3-subfamily and the subsequent depletion of LH 5′SS occurring early in fungal development, our data point to long-term coevolution between LUC7 subfamilies and 5′SS subclasses.
Conclusion
The study is consistent with a paradigm in which LUC7 proteins—more especially, LUC7L2/LUC7L and LUC7L3—are essential for splicing regulation in both plant and animal lineages. The U1:5′SS duplex is directly contacted by highly conserved portions of these proteins, which crosslink close to 5′SS and bind with U1 snRNP. Minigene studies confirm that certain 5′SS nucleotide characteristics, summed up by the 5′SS Balance score, are necessary for LUC7 proteins to affect pre-mRNA splicing. Domain-swapping tests demonstrate that specificity for 5′SS subclasses can be conferred by LUC7 structured regions alone. Therapeutic regulation of splicing, a critical mechanism in gene expression, is an attractive target. LUC7 proteins are a promising option for small molecule splicing regulators since they are associated with particular 5′SS sequence characteristics. The recognition of U1:5′SS RNA duplex structures is part of their regulation process, which may affect the specificity of small molecules that stabilize U1 snRNP:5′SS connections. Patients with AML who have monosomy seven or LUC7L2 mutations may benefit better from splicing therapies that target RH 5′SS recognition.
Article Source: Reference Paper | Reference Article.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.