The field of bioinformatics is witnessing a major transformation with the development of genome foundation models. In a recent study conducted by researchers from Northwestern University and Lawrence Berkeley National Laboratory, along with collaborators from multiple institutions, a groundbreaking model named GenomeOcean has been introduced. This 4-billion-parameter generative genome foundation model is trained on large-scale metagenomic assemblies, enabling it to represent diverse microbial species and generate biologically meaningful sequences.
The Challenge: Bias and Inefficiency in Genomic AI
Genomic AI models are ridden with issues today, and the first problem stems from the fact that they are trained heavily on reference genomes, which have been studied a lot. This leaves a whole myriad of microbes that go uncultured and are of low abundance, like those found in oceans, soils, and the human gut, which is known as the “rare biosphere.” Further, so-called “traditional” models do not bode well with the complexity of real-world metagenomic data, making them computationally sluggish.
GenomeOcean addresses these complications directly. Instead of using these traditional models, the team trained their model on DNA from the Tara Oceans expedition, metagenomes housed in Antarctic subglacial lakes, and even tropical soils, which amounts to about 645 billion base pairs. With these models, it becomes statistically possible to grasp the microbes that would otherwise remain undiscovered with the genome-centric methods.
The Innovation: Speed, Scale, and Biological Fidelity
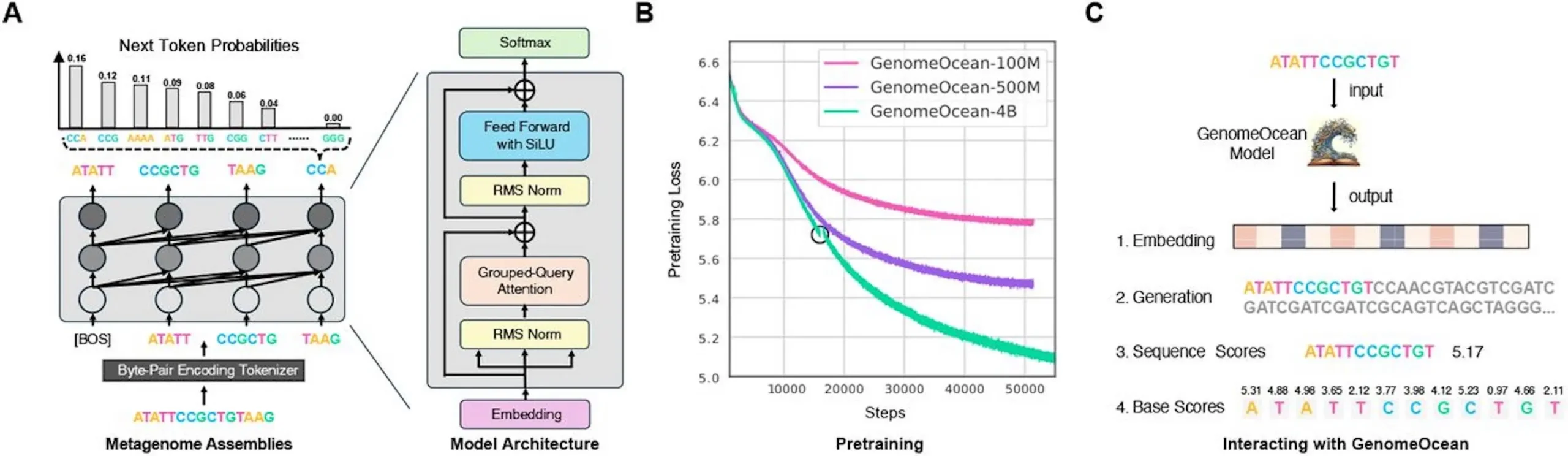
At its core, GenomeOcean is a 4-billion-parameter transformer model optimized for efficiency. The team implemented byte-pair encoding (BPE), a tokenization strategy borrowed from natural language processing, to compress DNA sequences into biologically meaningful chunks. This reduced sequence lengths by 5x, enabling GenomeOcean to generate DNA 150 times faster than comparable models like Evo-7B and GenSLMs-2.5B.
But speed is not the only one of its superpowers. Even when being trained on metagenomic contigs, which are far more fragmented (with a larger proportion being shorter than a single gene), GenomeOcean was able to synthesize full-length, functional proteins. When provided with the first part of a bacterial gene, it was able to append sequences in such a way that the result was three-dimensional structures almost indistinguishable from real proteins. Amazingly, it was even able to tell the difference between synonymous mutations (benign changes to the codon) and harmful mutations, demonstrating that it understands the “grammar” of protein-coding DNA.
Applications: From Drug Discovery to Ecosystem Insights
The implications are profound. In one experiment, the team fine-tuned GenomeOcean to specialize in biosynthetic gene clusters (BGCs)—genomic regions that produce antibiotics, anticancer compounds, and other bioactive molecules. The model was able to foster polyketide synthesis by multi-enzyme assembly lines, and it generated completely new biogeochemical domains. Additionally, it found new BGCs in already-known genomes, suggesting an additional supply for natural product breeding.
In addition to biotechnology, GenomeOcean performs well in basic microbial ecology. Its DNA embeddings clustered species as accurately as conventional tetra-nucleotide frequency techniques, even differentiating between close species like strains of E. coli and Salmonella. This decline in the efficiency of existing tools may transform metagenomic binning for especially low-abundance environments where they currently do not work well.
Safety and Accessibility
A standout feature is GenomeOcean’s self-awareness. When tasked with distinguishing its AI-generated sequences from natural DNA, it achieved 99% accuracy, outperforming other models. This built-in “detector” minimizes the risk of contaminating public databases with synthetic sequences—a critical safeguard for future applications.
The team has open-sourced GenomeOcean on GitHub alongside tools for BGC discovery and sequence validation. Training required immense computational firepower (14 days on 64 A100 GPUs), but the optimized inference code allows smaller labs to run the model on a single GPU.
A Step Towards the Future
GenomeOcean has taken a huge leap in the development of genome foundation models, as it introduces new ways of thinking and possesses unparalleled computational computational efficiency. It casts aside reference genome biases and integrates extensive metagenomic data, thus enabling new avenues in microbial studies, precision medicine, and synthetic biology.
As co-author Zhihan Zhou (Northwestern University) notes, “GenomeOcean is just the first dive into an ocean of genomic dark matter. With larger models and richer datasets, we’ll uncover evolutionary rules we’ve never imagined.”
For microbiologists, synthetic biologists, and AI researchers alike, GenomeOcean isn’t just a tool—it’s a lighthouse illuminating the hidden complexity of life’s code.
Article Source: Reference Paper | GenomeOcean is open source and publicly available on GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.