Protein pockets have been an integral part of drug discovery for several years and are also targeted during the design of novel molecules. Pockets refer to the parts of proteins where small molecules or ligands would be able to attach and mediate the desired effect. Creating the same, however, has been one of the daunting tasks in the hands of the scientists. A team of researchers from the University of Science and Technology of China and the Harvard Medical School—introduced PocketGen, an innovative tool that leverages deep learning to co-design protein pocket sequences and structures. This distinguishes itself from the traditional knowledge in the field and brings an elegant and effective solution to protein design and molecular targeting.
Why was this Study Conducted?
Traditional protein engineering often separates sequence generation and structural refinement into distinct processes. This reductive viewpoint may neglect the intricate relationships between the protein sequence and its three-dimensional organization. To overcome these issues, PocketGen was developed. The study aimed to improve the design of protein pockets—an essential step in optimizing interactions between that protein and other molecules for therapeutic purposes, thereby increasing its accuracy and efficiency by combining the two approaches.
Introducing PocketGen: A Unified Approach
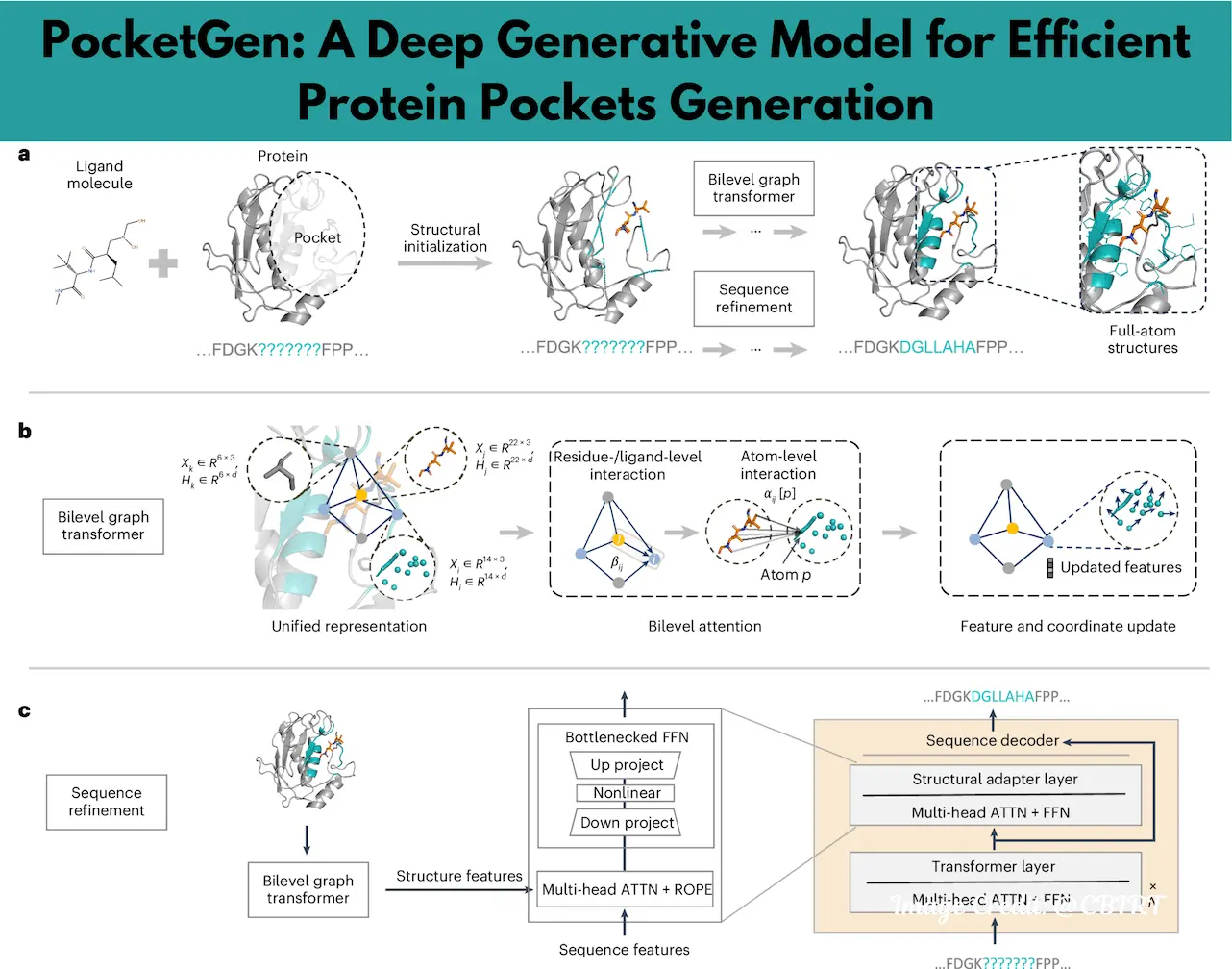
PocketGen takes an innovative approach by positioning protein pocket design as a generative task. This means that the design process is guided by the specific characteristics of the protein scaffold and the ligand molecule it needs to interact with. In contrast to other conventional approaches, PocketGen does not simply refine the existing structure of the pocket but also creates new ones by simultaneously considering sequence and structural elements.
To do so, the researchers used a bilevel graph transformer. This is an efficient model that is capable of staying on top of atomic interactions while resolving at the residue/ligand interaction level. Achieving this two-pronged approach enables PocketGen to interconnect the interactions of protein-ligand complexes with almost perfect fidelity. Furthermore, the integration of pretrained protein language models (pLMs), such as ESM, enhances the sequence prediction capabilities of the system. These models, augmented with structural adapters, enable PocketGen to generate pockets that are not only accurate but also biologically relevant.
How PocketGen Works
PocketGen is a sophisticated tool that co-designs the amino acid sequences and three-dimensional (3D) structures of protein pockets, specifically tailored to fit and bind with target ligand molecules. Advanced deep learning has been the backbone of pocket generation. Graph transformers and protein language models combined are what allow for an accurate and realistic functional pocket generation.
The first step involves the location of the protein pockets, which are defined as a group of residues located within some distance from the ligand molecule. Once the pocket region is determined, PocketGen generates its sequence and atomic structure. In addition, the system has implemented a recycling strategy whereby these outputs are gradually improved to meet biological and geometrical requirements.
The training process integrates three key objectives: optimizing the sequence, refining the coordinates, and ensuring structural integrity. Such an approach allows the generated pockets to fit properly onto their proteins as well as interact with their ligands.
Results that Speak for Themselves
PocketGen was tested comprehensively using two datasets: the cross-docked dataset, which contains 180,000 protein-ligand pairs, and the binding mode dataset, which includes a total of 41000 experimentally validated protein-ligand complexes. In these tests, PocketGen kept on demonstrating better performance than traditional methods and, even not the least, modern tools such as RFdiffusion and FAIR.
The most unique element of PocketGen is its ability to generate pockets with greater accuracy of ligand binding. With this nature, it becomes a valuable tool in the drug discovery process where the accuracy of molecular interactions is very critical. Thanks to PocketGen, which increases the possibility of creating successful therapeutics by designing pockets that are closest to the ligands.
Applications and Future Potential
The application of the features of PocketGen goes way beyond the boundaries of drug development. In synthetic biology, for example, it may be applied to modify proteins in a way that they gain fundamentally new functions. In therapeutic design, it is a way to pursue a better molecular interaction, leading to enhanced therapeutic performance and diminished toxicity. Even though it has proved to be successful, the researchers believe that there are ways that the design can be improved. They suggest that logic in chiral properties would be influenced positively by including SE(3) equivariant models as a way to refine PocketGen. Additionally, expanding the training datasets to encompass a broader range of protein-ligand interactions would make the tool even more robust.
Conclusion: A Step Forward in Protein Design
PocketGen demonstrates a major advancement in protein design as it integrates deep learning, structural biology, and computational efficiency into one system. Allowing for the fast and accurate co-design of drugable protein pockets removes a major hurdle in the drug development race and raises the bar for more progress to come. Looking ahead, such technologies as PocketGen will transform the process of drug design into a faster and more accurate one, eventually enabling researchers around the world to use them. Not only does this development showcase the capabilities of AI in biology, but it also strengthens the position of interdisciplinary solutions to multidimensional scientific challenges.
Article Source: Reference Paper | The source code for this study is made freely available by researchers on GitHub and Zenodo.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.