Federated machine learning enables the largest brain tumor study ever conducted without sharing patient data by allowing researchers to securely analyze distributed datasets while maintaining data privacy. The breakthrough research from Penn Medicine and Intel Corporation published in Nature Communications has the potential to revolutionize medical research and improve our understanding of brain tumors. It also provides a secure way for organizations to collaborate on large-scale studies while protecting patient data.
Ever wondered how your Gboard (Google keyboards) provides next-word suggestions or query recommendations based on what you are typing? We probably know that it has something to do with machine learning (ML) or deep learning, and if you know enough about machine learning, you would know that for an ML model to make predictions, it has to be trained on some data. An ML model as widespread as the Gboard needs to have good generalizability, which is acquired through training on diverse real-world data, and copious amounts of it too. And if you think about it, it’s not that hard for tech giants such as Google to grab their user’s data to train their models. But could you just imagine the lawsuits zipping their way to Google if they were to invade user privacy? If so, how come there is no noose around Google’s neck yet? Well apparently, they escaped public execution the same way the researchers at the University of Pennsylvania outsmarted the stalemates in healthcare data sharing – through Federated Learning.
Nothing shouts’ confidential’ more than healthcare data, and with good reason. But in this day and age where a lot of clinical decisions could be assisted by computerized prediction models holding data confidential could also mean a big dent in what otherwise could have been a highly robust model for making accurate clinical predictions. The regulatory policies across various countries for protecting patient information, such as HIPPA in the US, DISHA in India, and GDPA in the European Union, make it almost impossible to collaborate across the globe. Especially in cases of rare diseases (present in less than 10 out of 100,000 people), a lack of effective global collaborations implies a lack of sufficient data. So, how could one overcome this impasse? Or better yet, could we ever overcome this impasse? And you know what, it seems that we have.
Federated Learning
Rather than giving medical data the center stage and all the limelight, how about we allow the data to be the background prop that it loves to be? Imagine a hypothetical situation: seeds are distributed from a store to a certain number of farmers, they plant it, water it, and give it the necessary nutrients to mature and grow, they then collect all the yield and send it back to the store. Not the most ideal example, I understand, and I am definitely against farmer exploitation, but the scenario could be substituted as follows: seeds can be an initial prediction model trained with the amount of data available at hand, which is then distributed to different sites across the globe to nurture and update the model through training on local data. The yield, which is the updated model is sent back to the store or the central location in the form of weights and biases of the model updated to accommodate the local data.
This system provides the benefits of training on large amounts of data without actually centralizing them at a single location. The updated information from different sites is aggregated together at the central location to build an improved model. This step is iterated with the improved model being deployed again to the next set of sites to be trained on the data held at those locations. This iteration continues until a predefined condition is attained. Google’s Gboard was the very first real-world application of Federated Learning that enabled them to provide better recommendations and predictions to users without invading their privacy through data collection. And this is also how a glioblastoma boundary classifying model was built through data from 71 sites across six continents, surpassing all the potholes of healthcare data gathering.
Federated Learning for Glioblastoma Boundary Prediction
Federated learning is still a budding field, and even though it has obvious potential in medical research, little of it is currently explored in the said field. But a team of scientists at the University of Pennsylvania sought to change it and ended up building the largest real-life federation learning network for healthcare data by garnering collaboration from across 71 sites and utilizing 6,314 multi-parametric MRI (mpMRI) scans of glioblastoma patients for accurate compartmentalization of glioblastoma. The study by itself generated the largest glioblastoma dataset to date to exist in literature.
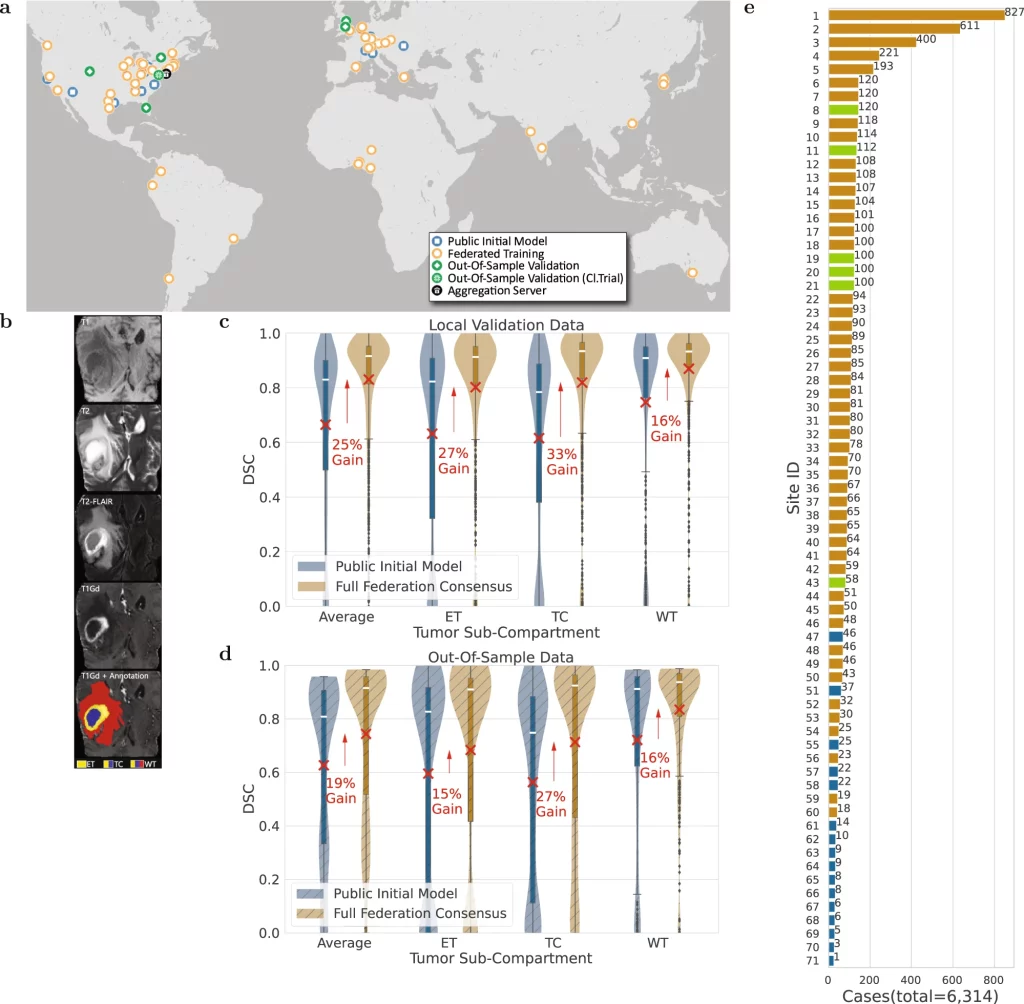
Image Source: https://doi.org/10.1038/s41467-022-33407-5
Glioblastoma, though the most common type of malignant brain tumor, is considered a rare disease due to its occurrence being around 3 in 100,000 people, which again stresses the need for information gathering. The glioblastoma can be separated into Enhancing Tumor(ET), Tumor Core(TC), and Whole Tumor(WT) sections. The ability to make accurate predictions for delineating these subsections from mpMRI scans could allow better quantification and assessment of the disease and assist clinicians in making better decisions for improving patient treatment and survival.
A ‘public initial model’ was developed at the centralized server in the University of Pennsylvania from 231 cases obtained from 16 sites gathered through the BraTS 2020 challenge based on the 3D-ResUNet network. This model was distributed to 35 sites where it was trained on a total of 2471 patient cases. The updated weights and biases returned to the centralized server were aggregated by the federated averaging method to build the ‘primary consensus model.’ The federated learning concluded with the ‘final consensus model’ by distributing the primary consensus model to the remaining sites and subsequent model updating. At all the sites, the data was divided in an 80:20 fashion, where 20% of the data was dedicated to the validation set. The entire data was also partitioned to leave out six untrained sites with 590 patient cases and 332 cases from the original dataset at the centralized server to be used as untrained out-of-sample data to validate the model further. The model performance was assessed by a Dice similarity coefficient and the model showed an average improvement gain of 25% for validation sets and 19% for out-of-sample datasets between the public initial model and the final consensus model. As a side note, the improvement of the delineating accuracy in both cases was higher for the TC section (33% and 27% for validation and out-of-sample datasets, respectively), which is the surgically actionable compartment and is hence of greater clinical significance.
Takeaways
The increased accuracy and generalizability in the compartment delineation were gained solely through the use of additional diverse data that emphasizes the need for more collaborations that allow for the exchange of information. From assessing the eligibility of the collaborating sites to coordinating the said collaborations to proper and harmonized pre-processing of the data, building a federation learning model is by no means a cakewalk. But if there is something we can take away from this study, is that it is worth the effort? Being one of the first to employ a federated model at a scale as large and complex as this, the authors have compiled invaluable insights into its proper governance. They have set the foundation for large-scale federated learning in medical research, hoping that it would have a positive impact on the treatment of glioblastoma and that it would lead to more successful federated learning networks in the future. But more importantly, they hope that this work could convey the effectiveness of large-scale federated learning and encourage more multi-site collaborations.
Article Source: Reference Paper | Reference Article
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Catherene Tomy is a consulting Content Writing Intern at the Centre of Bioinformatics Research and Technology (CBIRT). She has a master’s degree in Molecular Medicine from Amrita University with research experience in the fields of bioinformatics, cell biology, and molecular biology. She loves to pull apart complex concepts and weave a story around them.
.







