Drug discovery and protein engineering are some of the most challenging aspects of cutting-edge scientific research. It is also a field that holds great promise in addressing some of the burning issues in global healthcare. Unfortunately, the intricacies of protein molecule interactions, dynamics, and the effects of mutations are exceedingly painstaking and slow. Researchers from the University of Montreal introduced NRGSuite-Qt, a PyMOL plugin designed to simplify and accelerate high-throughput screening, molecular docking, normal-mode analysis, and the study of protein-protein interactions. By integrating a suite of essential tools in a single, user-friendly interface, NRGSuite-Qt provides an accessible platform for researchers to conduct complex analyses with reduced computational resources.
What is NRGSuite-Qt?
NRGSuite-Qt is a user-friendly plugin for PyMOL, a well-known molecular visualization program. This bioinformatics package was developed to aid the researchers in all aspects of drug design and protein engineering, including virtual screening, docking, mutation analysis, and binding-site contour matching. One of the main benefits of NRGSuite-Qt is that it integrates numerous methods in a single system, which makes it easier for researchers to solve complex scientific problems faster and more effectively.
At its heart, NRGSuite-Qt employs a coarse-grained method, which enables it to deliver output at a lower computational cost. This makes it perfect for multi-sample applications. The conceptual framework’s ease of use also helps lower the barriers to utilization and allows researchers of diverse backgrounds to be able to fully use its features.
Key Features and Capabilities of NRGSuite-Qt
Advanced users of molecular analysis processes for drug discovery, protein engineering, and structural bioinformatics will find NRGSuite-Qt to be a remarkably effective and user-friendly platform. One of the highlighted features is high-throughput virtual screening, where researchers can evaluate large compound libraries against target proteins, which makes it perfect for drug repurposing and primary stages of drug discovery. The platform further offers the capability of molecular docking, simulating small molecules, such as drugs, to specific protein targets for binding. This assists in understanding their effectiveness and interaction. In addition to those features, NRGSuite-Qt also uses normal mode analysis (NMA), which helps examine the motion and shape changes of proteins, which is important in knowing the proteins’ strengths and actions. This is very helpful when observing large proteins or protein complexes that may experience a lot of change due to surrounding factors.
NRGSuite-Qt is focused on studying protein-protein interactions (PPIs) and searching for similarities in the binding sites of different proteins. From the points of contact of proteins, scientists can determine important residues from these complex proteins that assist in modifying biologically active compounds and, by this means, aid in drug design for novel targets. The system allows extended drug repurposing by comparing binding sites in different families of proteins and reveals those compounds that can interact with similar regions in other proteins. NRGSuite-Qt offers a variety of tools for molecular docking, PPI studies, and binding-site analysis, thus the system takes into consideration all structural features omega which determines targets of interest. Such integration of these methods can be performed more effectively via one program. This control has increased the application of NRGSuite-Qt in drug development and protein engineering because it has become easier to use.
Case Studies: Real-World Applications
To showcase the efficacy of NRGSuite-Qt, the developers put the system through several practical exercises:
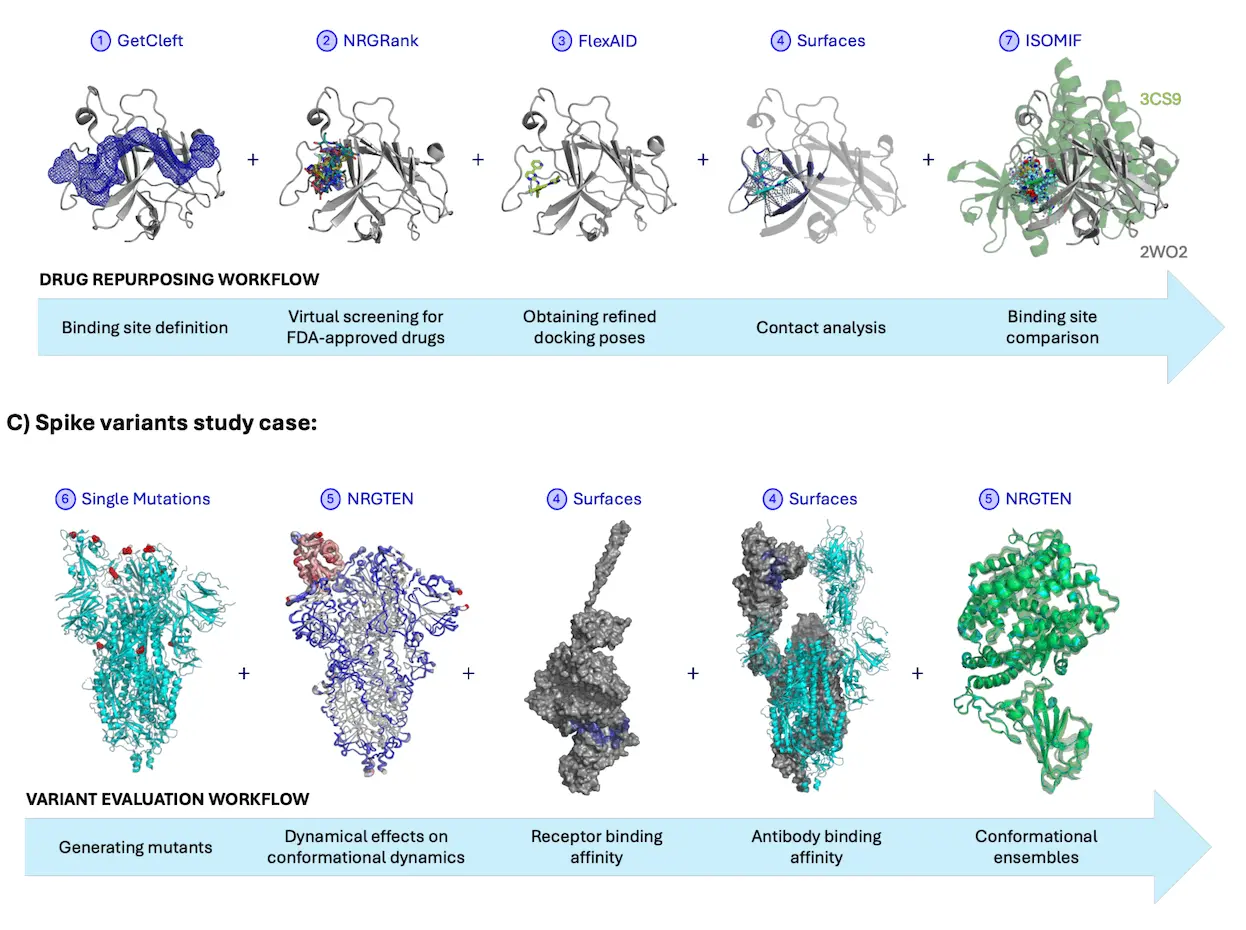
EphA4 Drug Repurposing
It was a study that sought to identify which of the FDA-approved drugs could inhibit the EphA4 receptor involved in Alzheimer’s disease, and the researchers were able to replicate it. NRGSuite-Qt was able to successfully locate Nilotinib as a promising inhibitor for this receptor and proved it was capable of predicting docking poses and binding interactions within the target site.
SARS-CoV-2 Spike Protein Variant Evaluation
Investigators also replicated the data on the mutations of the SARS-CoV-2 virus. With the variant in the spike protein, NRGSuite-Qt demonstrated how certain mutations altered protein flexibility and binding to the ACE2 receptor, which now shed light on how the virus may enhance its infectivity and evade immune response.
Conclusion
NRGSuite-Qt revolutionizes drug discovery and protein engineering by providing a literal all-in-one software tool for molecular modeling and high-throughput screening. It allows users to dock proteins and run normal-mode analysis, evaluate mutations, and perform protein-protein interaction studies without switching from one tool to another! This is a crucial leap forward in the field.
NRGSuite-Qt promises to set new records in drug discovery, protein engineering, and other scientific disciplines. From experienced bioinformaticians to novices planning to dip their toes into the world of molecular dynamics, everyone will find an elegant yet powerful environment within this plugin for solving difficult challenges in medicine and other fields.
Article Source: Reference Paper | The Installation guide and tutorial are available on the Website.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.