Scientists at UC Santa Cruz, Google, the National Institutes of Health, and partner institutes have developed DeepSomatic, a deep-learning tool that detects cancer-related DNA alterations (somatic variations) from both short-read and long-read sequencing. Unlike other tools, it can operate with whole-genome, exome, tumor-normal, tumor-only, and FFPE samples. The team generated and publicly released five matched tumor-normal datasets sequenced on the Illumina, PacBio, and Oxford Nanopore platforms. DeepSomatic surpasses existing approaches in all three technologies, particularly indel detection.
Overview
Cancer is primarily a genomic disease characterized by somatic DNA mutations. Detecting these is critical for providing accurate, personalized treatment. Somatic variant calling compares tumor DNA to matched normal samples; however, widely used tools such as Strelka2 and MuTect2 use short-read sequencing, which limits the detection of complex variants, mutations in repetitive regions, and low-frequency tumor variants in samples containing normal cells. Long-read sequencing methods, including ONT and PacBio HiFi, serve to address these issues by creating extremely accurate long reads that increase the detection of complicated and rare mutations. However, progress remains restricted by a lack of large, actual somatic training datasets, forcing many variant callers to rely on simulated data, which may not adequately represent the true diversity of cancer mutations.
What is DeepSomatic?
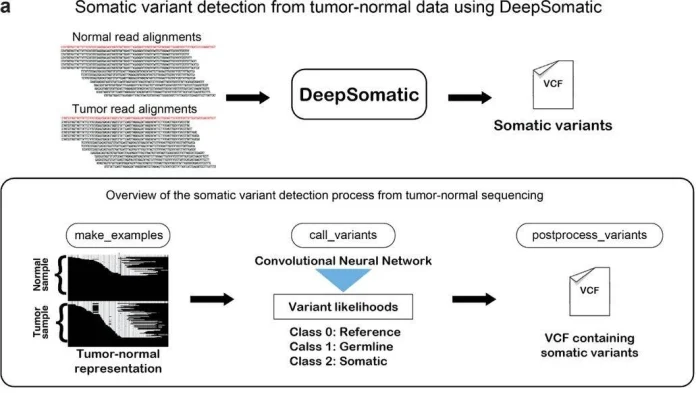
DeepSomatic is a tool to detect somatic mutations in cancer that works with both short-read and long-read DNA sequencing. It is based on DeepVariant, a deep learning method originally designed to detect germline mutations. DeepSomatic works by combining DNA readings from tumor and normal samples, as well as the reference genome, to create visual representations known as pileup images, which are then analyzed using a convolutional neural network (CNN) to determine which changes are actual mutations.
Unlike DeepVariant, to enhance its accuracy, the developers combined five different cancer cell lines (three breast cancer: HCC1395, HCC1937, HCC1954, and two lung cancer: H1437 and H2009) with their corresponding normal cell lines. To ensure accuracy, they sequenced each pair using three technologies: Illumina (short reads), PacBio HiFi, and Oxford Nanopore (long reads), all from the same DNA sample. The sequencing produced high coverage for both tumor and normal DNA, and the long reads were extremely long (tens of kilobases), making it easier to find complicated mutations. They built a high-confidence set of genuine cancer mutations by including only changes found by at least two separate sequencing models and deleting unreliable genomic areas.
They verified the method’s accuracy by demonstrating 93% agreement with an official benchmark dataset that also indicated that each cancer cell line has unique mutation patterns and variant frequencies. Furthermore, it was also effectively modified for Formalin-Fixed Paraffin-Embedded (FFPE) samples (hospital tissue samples with DNA damage) and whole-exome sequencing (WES), which sequences only genes. Across these hard data types, DeepSomatic made fewer errors and recognized mutations accurately than existing techniques such as ClairS and Strelka2, which is why training on multiple cancers improves DeepSomatic’s performance to handle realistic clinical circumstances. As a result, it can accurately detect somatic mutations across many sequencing technologies and outperforms prior approaches.
How DeepSomatic was Built and Tested
Five cancer cell lines and their matched normal counterparts were cultivated, enumerated, and flash-frozen before being extracted for high-quality DNA. DNA was sequenced with three technologies: Illumina (short reads), PacBio HiFi, and Oxford Nanopore (ONT, long reads). Sequencing reads were aligned to the human reference genome (hg38) with BWA-MEM2, pbmm2, and minimap2, and metrics such as alignment identity, N50, and coverage were determined. DeepSomatic detected somatic variants for tumor-only analysis by comparing tumor reads to population allele frequencies and then filtering using normal variant panels.
Combining data from several models and technologies provided high-confidence variant sets, while BED files were used to define appropriate genomic regions. DeepSomatic’s performance was compared to tools like Strelka2, ClairS, MuTect2, and SomaticSniper, utilizing SEQC2 data. Titration BAMs simulated contamination to determine reliability, and SigProfiler was used to investigate patterns of single-base substitutions and indels, offering insight into tumor-specific mutational processes.
Conclusion
DeepSomatic outperforms current somatic variant callers on both short-read and long-read sequencing systems, particularly for recognizing difficult indels. This makes it a more reliable tool than other tools such as Strelka2 and ClairS. Furthermore, the authors have made all of the sequencing data and benchmarking datasets openly available without restrictions, helping the scientific community generate and evaluate somatic variant-calling methods.
Article Source: Reference Paper | Code availability: GitHub
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Jainab Shaikh is a postgraduate in Biotechnology with a strong interest in understanding how research translates into real-world innovation. Her areas of focus include biosensors, bioinformatics, and sustainable biotechnological applications. She is passionate about exploring recent scientific advancements and communicating them through clear, engaging, and accessible content. Her work particularly emphasizes research-driven narratives in healthcare, biotechnology, skincare science, and emerging life science innovations.