Hospital for Special Surgery (HSS) researchers in New York City have developed a computer vision tool that effectively identifies rheumatoid arthritis (RA) from osteoarthritis (OA) in joint tissue collected from patients who had total knee replacements (TKR).
The findings show that the new computer vision model with improved proficiency to differentiate rheumatoid arthritis from osteoarthritis will aid in the short-term improvement of research procedures and the long-term optimization of patient care. The results were presented at the 2022 European Alliance of Rheumatology Congress (EULAR) in the first week of June.
Computer vision nuclei counting can revolutionize how inflammation is assessed in synovial and other tissue biopsies by providing an accurate, automatic, cost-effective, and quantitative method that can be widely distributed and used. Automated nuclei detection is a critical step in generating consistent and repeatable assessments of synovial inflammation, which are needed to produce guidelines for incorporating histological results in RA diagnosis and therapy decisions.
We know there are many more immune cells present in the synovium, or joint tissue, of patients with RA, compared to those with OA. But precisely how many more have not been clear.
Bella Mehta, MBBS, MS, a rheumatologist at HSS and lead author of the study.
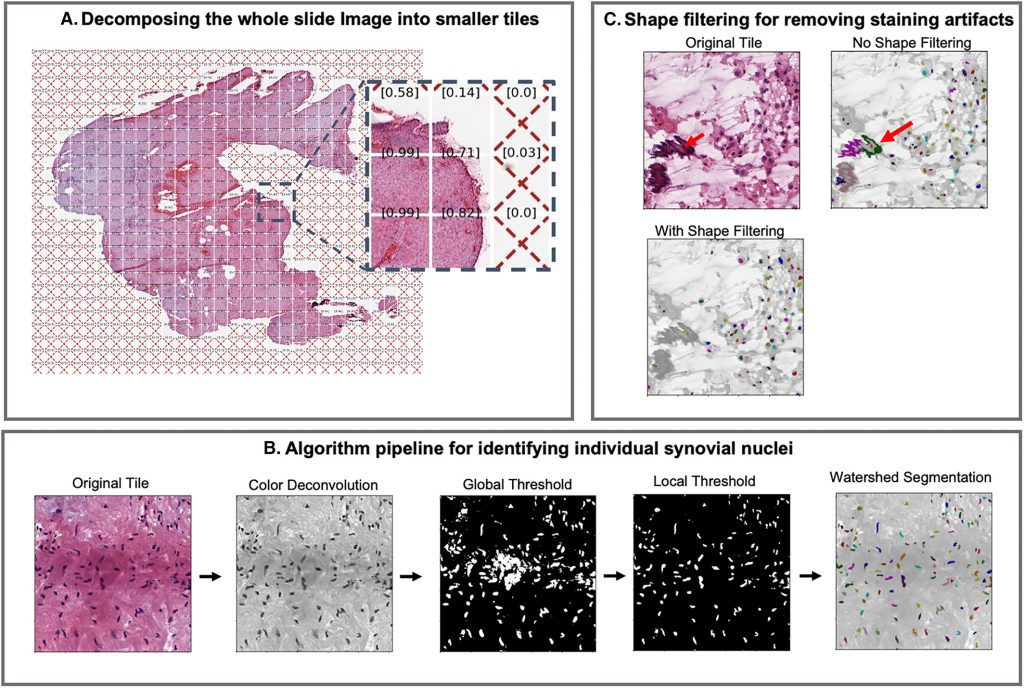
Image Source: Rheumatoid Arthritis Synovial Inflammation Quantification Using Computer Vision.
Total knee replacement is frequently the only treatment choice for individuals with extensive knee joint injury. Given that RA is a systemic, inflammatory illness that can harm the eyes or the lining around the heart, while OA only affects the joints, determining which disease caused the joint damage is critical for guiding treatment regimens. For instance, in a recent study conducted by HSS researchers, assessments from two highly experienced pathologists analyzing the infiltration of one type of immune cell known as lymphocytes on the same slides only agreed 67 percent of the time.
Pathologists typically assess images of synovium to determine the extent of inflammation using a combination of approaches, including assigning the level of immune cell infiltration on a scale from 0 to 4. However, these methods are imperfect.
Dana Orange, MD, MS, a rheumatologist at HSS, assistant professor at Rockefeller University and senior author of the study.
Dr. Orange and Dr. Mehta and their collaborators from HSS and other institutions developed and validated a computer vision method that counts tens of thousands of cell nuclei in whole-slide synovium pictures. They employed a computer vision technique to evaluate cell density and measured 14 different pathologist-scored features in synovium from 60 patients with RA and 147 patients with OA who underwent TKR.
Using traditional computer vision approaches, the algorithm identifies synovial nuclei based on color, shape, and size. Other histological features and artifacts, such as excessive staining that looks like nuclei, can be mistakenly classified as nuclei. Nuclei in dense clusters, particularly those with contiguous or overlapping nuclei, might be mislabeled as a single nucleus. As shown in the validation investigation, the number of nuclei vastly outnumbers the number of incorrectly recognized items. As a result, while overall slide image statistics are unaffected by this type of mistake, local data on smaller areas may be.
The researchers discovered substantial differences in synovium characteristics between RA and OA. Compared to the OA samples, the RA samples had higher cell density, fewer mast cells (a kind of white blood cell), and fewer indications of fibrosis or scarring. When the 14 pathologist-scored features were used alone, the probability of correctly distinguishing between RA and OA in synovium was 85 percent, 88 percent when the computer’s cell density score was used alone, and 91 percent when the pathologists’ scores and the computer’s cell density calculation were combined. The researchers came up with a cutoff threshold for separating RA from OA, deciding that synovium with more than 3,400 cells per mm2 should be categorized as RA.
According to Dr. Orange, the innovation is not yet suitable for clinical usage, but it has the potential to help pathologists in the future. For the time being, the researchers regard it as a valuable research tool because it delivers an accurate and 100 percent reproducible score of inflammation, and they look forward to further developing it.
The researchers believe that in the future, computer vision might be trained to extract additional information from tissue samples, such as the sorts of cells present and whether they are near enough together to communicate with one another. Clinicians may be able to determine more precisely which cells are causing tissue damage as a result of this more granular examination and modify treatments accordingly.
Story Source: Guan S, Mehta B, Slater D, et al. Rheumatoid Arthritis Synovial Inflammation Quantification Using Computer Vision. ACR Open Rheumatol. 2022;4(4):322-331. doi:10.1002/acr2.11381
Orange DE, Agius P, DiCarlo EF, et al. Identification of Three Rheumatoid Arthritis Disease Subtypes by Machine Learning Integration of Synovial Histologic Features and RNA Sequencing Data. Arthritis Rheumatol. 2018;70(5):690-701. doi:10.1002/art.40428
https://news.hss.edu/computer-vision-tool-improves-the-ability-to-distinguish-rheumatoid-arthritis-from-osteoarthritis-in-damaged-joint-tissue/
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.