Researchers at IBBM (Instituto de Biotecnología y Biología Molecular) and NOVABIOMA Biomanufacturing FlexCo introduced ColabPCR, a Google Colab-based Python notebook created along with collaborators in Austria and Canada. It simplifies PCR primer design by combining Primer3 for primer selection, BLASTn for off-target detection, and restriction site insertion in a single platform. Validated on carrot genomes, ColabPCR increased PCR success rates, particularly when off-target primers were removed, making primer design simpler and more dependable.
Primer Design in PCR: Principles and Existing Computational Approaches
In Molecular Biology, the polymerase chain reaction (PCR) is a common technique used to amplify a specific sequence of DNA, and the amplification depends on well-designed primers. A good primer must have balanced GC content and a suitable melting temperature, as the primer must not form a hairpin model or bind to itself. At present, we have several software programs like Primer3, Primer-BLAST, QuantPrime, and PRIMEGENS to assist in creating primers.
These programs work by taking a target sequence, and users specify parameters like primer length, melting point, and product size, to create the desired primer candidate. This software uses thermodynamic models to estimate how well primers will bind to the given target sequence. Further, an additional step is to conduct a BLAST search to eliminate primers that closely match the untargeted genomic sequence in DNA.
Even with these precautions, off-target primer binding may happen simply because primers can bind to several repeating sequences similar to our target DNA sequence, or due to mismatches, which leads to non-specific amplification, misleading the results, as well as low PCR efficiency.
The existing methods focus on picking primers that are meant to be specific rather than providing details regarding the relevant non-specific binding activity, which is important for troubleshooting PCR failures or refining primer design.
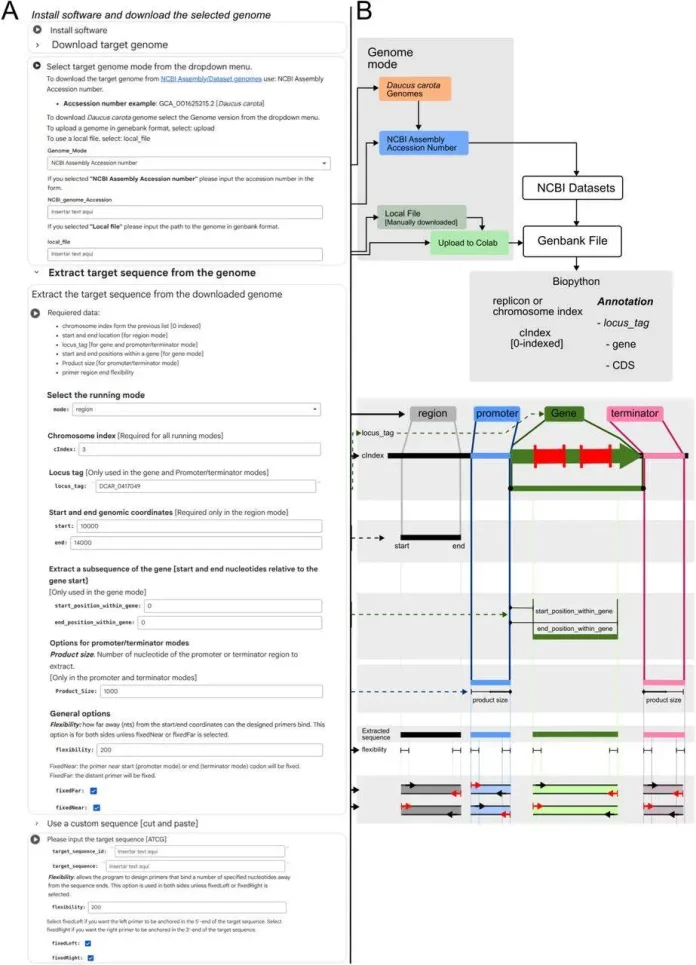
An Insight into Google CoLabPCR and its workflow
CoLabPCR is a python notebook implemented on Google Colab, it helps user to design primer more accurately than the existing tools especially for cloning experiments, it serves as a one stop tool which enables user to design accurate primer also gives insight into the non specific binding activity of the primer as well as the prediction for using the specific restriction enzyme to avoid cutting inside of the target DNA sequence.
To use the CoLab the first step is to create the work environment by installing the software and selecting the genome (eg; Daucus carota, Ergo Bioscience SAS proprietary cell lines), through the NCBI Assembly Accession number or by locally uploading the file (genebank file), then extracting the targeted sequence from the genome by selecting the criteria eg chromosome number, start and end point for genomic coordinate, promoter, terminator.
The “flexibility” option allows primers to bind a few bases inside the target region rather than strictly at the ends. The FixNear and FixFar options can lock primer positions while allowing only primer length to change. FixNear binds the primer on the start-codon side in region, gene, and promoter modes, and on the stop-codon side in terminator mode, always following the coding-strand direction.
Next, restriction enzyme sites are identified within the target sequence, and 5′ oligonucleotides and restriction sites are added to the forward and reverse primers (The forward (fwd) primer binds on the 5′ end of the target sequence. The reverse (rv) primer binds in the 3′ end of the target sequence). Primer3 is then used to create primers with the ideal size, GC content, and melting temperature. Finally, BLASTn-based virtual PCR detects off-target amplification.
In batch 1, CoLabPCR was used to create primers and the researchers did not test off-target binding with virtual PCR (only choose the primer combinations with the best Primer3 design scores), the primers were directly tested in experimental PCR. Out of all reactions, 62% resulted in the right PCR product. This demonstrated that high-scoring primers operate frequently, but not always, as some still bind in undesired locations.
In batch 2, after constructing primers, they performed virtual PCR (BLASTn) to eliminate primers that could bind elsewhere in the genome or had mismatches. When these were tested in the lab, the success rate rose to 71%. Significantly, some targets that failed in Batch 1 worked in Batch 2, demonstrating that checking for off-target binding is quite useful.
In batch 3, they used ColabPCR on a cloning project. Primers were created for the promoter and terminator regions, and restriction enzyme sites were inserted at the 5′ ends to allow the PCR products to be cloned. These primers were again tested using virtual PCR. This time, 74% of reactions successfully amplified the expected product (83 of 112). After cloning and sequencing, 68% of the constructions were confirmed as valid. This demonstrated that ColabPCR performs effectively even in high-throughput cloning efforts.
Conclusion
ColabPCR is a Python notebook that automates and improves primer design in molecular biology. It runs on Google Colab and combines genome extraction, Primer3-based primer building, restriction site testing, and whole-genome virtual PCR into a single, easily accessible procedure. One of its advantages is its accurate reporting of off-target binding sites and prediction of real PCR products based on primer orientation and distance; these features make primer selection easier, more precise, and more consistent than existing software.
Article Source: Reference Paper | Availability: GitHub and Jupyter notebook.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.